8TO1
 
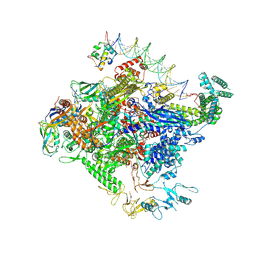 | | Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter | | 分子名称: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Darst, S.A, Saecker, R.M, Mueller, A.U. | | 登録日 | 2023-08-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
6NWL
 
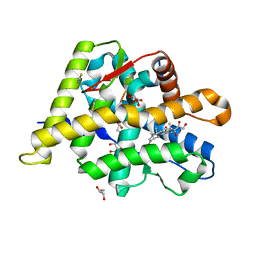 | | Structure of the Ancestral Glucocorticoid Receptor 2 ligand binding domain in complex with hydrocortisone and PGC1a coregulator fragment | | 分子名称: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | 著者 | Liu, X, Ortlund, E.A. | | 登録日 | 2019-02-06 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.595 Å) | | 主引用文献 | First High-Resolution Crystal Structures of the Glucocorticoid Receptor Ligand-Binding Domain-Peroxisome Proliferator-ActivatedgammaCoactivator 1-alphaComplex with Endogenous and Synthetic Glucocorticoids.
Mol.Pharmacol., 96, 2019
|
|
8XA9
 
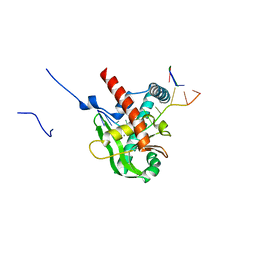 | | Human MGME1 in complex with 5'-overhang DNA | | 分子名称: | CALCIUM ION, DNA (11-MER), DNA (18-MER), ... | | 著者 | Wu, C.C, Mao, E.Y.C. | | 登録日 | 2023-12-03 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural basis of how MGME1 processes DNA 5' ends to maintain mitochondrial genome integrity.
Nucleic Acids Res., 52, 2024
|
|
6NWK
 
 | |
5LPX
 
 | | Crystal structure of PKC phosphorylation-mimicking mutant (S26E) Annexin A2 | | 分子名称: | Annexin A2, CALCIUM ION, GLYCEROL | | 著者 | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | 登録日 | 2016-08-15 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
8V07
 
 | |
5LQ2
 
 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 3.4 A resolution | | 分子名称: | Annexin A2, CALCIUM ION | | 著者 | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | 登録日 | 2016-08-15 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5LM7
 
 | | Crystal structure of the lambda N-Nus factor complex | | 分子名称: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | 著者 | Said, N, Santos, K, Weber, G, Wahl, M.C. | | 登録日 | 2016-07-29 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
5LM9
 
 | | Structure of E. coli NusA | | 分子名称: | MAGNESIUM ION, SULFATE ION, Transcription termination/antitermination protein NusA | | 著者 | Said, N, Weber, G, Santos, K, Wahl, M.C. | | 登録日 | 2016-07-29 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.143 Å) | | 主引用文献 | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
5LPU
 
 | | Crystal structure of Annexin A2 complexed with S100A4 | | 分子名称: | Annexin A2, CALCIUM ION, GLYCEROL, ... | | 著者 | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | 登録日 | 2016-08-15 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5LQ0
 
 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 2.9 A resolution | | 分子名称: | Annexin A2, CALCIUM ION | | 著者 | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | 登録日 | 2016-08-15 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
8V06
 
 | |
8V08
 
 | |
8V05
 
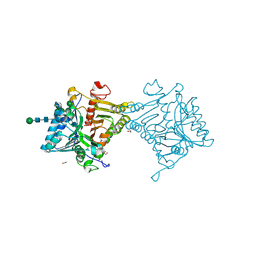 | | Crystal structure of mouse PLD3 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2023-11-17 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
7NYX
 
 | |
7NYY
 
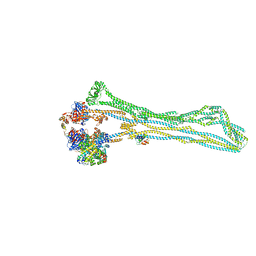 | | Cryo-EM structure of the MukBEF monomer | | 分子名称: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | 著者 | Buermann, F, Lowe, J. | | 登録日 | 2021-03-23 | | 公開日 | 2021-07-07 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ4
 
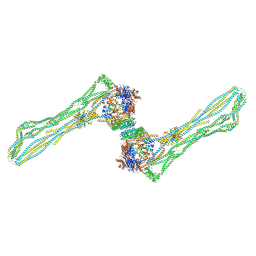 | | Cryo-EM structure of the MukBEF dimer | | 分子名称: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | 著者 | Buermann, F, Lowe, J. | | 登録日 | 2021-03-23 | | 公開日 | 2021-07-07 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (13 Å) | | 主引用文献 | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYW
 
 | |
7NZ2
 
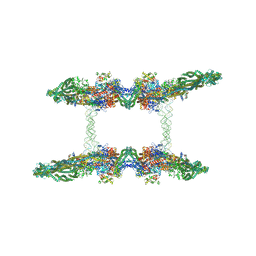 | | Cryo-EM structure of the MukBEF-MatP-DNA tetrad | | 分子名称: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | 著者 | Buermann, F, Lowe, J. | | 登録日 | 2021-03-23 | | 公開日 | 2021-07-07 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYZ
 
 | |
7NZ0
 
 | |
7NZ3
 
 | |
1EJ9
 
 | | CRYSTAL STRUCTURE OF HUMAN TOPOISOMERASE I DNA COMPLEX | | 分子名称: | DNA (5'-D(*C*AP*AP*AP*AP*AP*GP*AP*CP*TP*CP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*C*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*GP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), DNA TOPOISOMERASE I | | 著者 | Redinbo, M.R, Champoux, J.J, Hol, W.G. | | 登録日 | 2000-03-01 | | 公開日 | 2000-08-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA.
Biochemistry, 39, 2000
|
|
1DML
 
 | | CRYSTAL STRUCTURE OF HERPES SIMPLEX UL42 BOUND TO THE C-TERMINUS OF HSV POL | | 分子名称: | DNA POLYMERASE, DNA POLYMERASE PROCESSIVITY FACTOR | | 著者 | Zuccola, H.J, Filman, D.J, Coen, D.M, Hogle, J.M. | | 登録日 | 1999-12-14 | | 公開日 | 2000-03-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase.
Mol.Cell, 5, 2000
|
|
1FLZ
 
 | | URACIL DNA GLYCOSYLASE WITH UAAP | | 分子名称: | URACIL, URACIL-DNA GLYCOSYLASE | | 著者 | Werner, R.M, Jiang, Y.L, Gordley, R.G, Jagadeesh, G.J, Ladner, J.E, Xiao, G, Tordova, M, Gilliland, G.L, Stivers, J.T. | | 登録日 | 2000-08-15 | | 公開日 | 2001-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase.
Biochemistry, 39, 2000
|
|
