4B6E
 
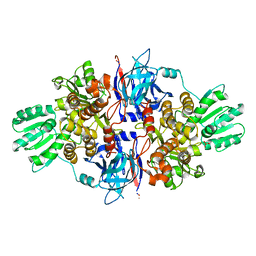 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | 分子名称: | 1H-indazol-7-amine, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | 著者 | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | 登録日 | 2012-08-09 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
4AJZ
 
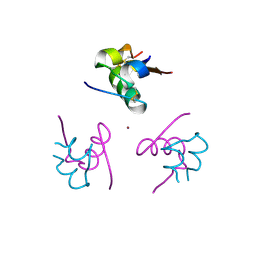 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | 分子名称: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | 著者 | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | 登録日 | 2012-02-21 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
2XFV
 
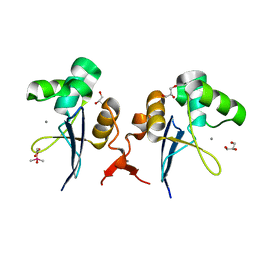 | | Structure of the amino-terminal domain from the cell-cycle regulator Swi6 | | 分子名称: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | 著者 | Smerdon, S.J, Goldstone, D.C, Taylor, I.A. | | 登録日 | 2010-05-27 | | 公開日 | 2010-06-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the Amino-Terminal Domain from the Cell-Cycle Regulator Swi6S
Proteins, 78, 2010
|
|
2D1C
 
 | |
2CNM
 
 | | RimI - Ribosomal S18 N-alpha-protein acetyltransferase in complex with a bisubstrate inhibitor (Cterm-Arg-Arg-Phe-Tyr-Arg-Ala-N-alpha- acetyl-S-CoA). | | 分子名称: | 30S RIBOSOMAL PROTEIN S18, COENZYME A, MODIFICATION OF 30S RIBOSOMAL SUBUNIT PROTEIN S18 | | 著者 | Vetting, M.W, Yu, M, Bareich, D.C, Blanchard, J.S. | | 登録日 | 2006-05-22 | | 公開日 | 2007-05-22 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Rimi from Salmonella Typhimurium Lt2, the Gnat Responsible for N{Alpha}- Acetylation of Ribosomal Protein S18.
Protein Sci., 17, 2008
|
|
2VK0
 
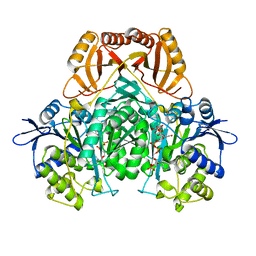
 | | Crystal structure form ultalente insulin microcrystals | | 分子名称: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, INSULIN A CHAIN, INSULIN B CHAIN, ... | | 著者 | Wagner, A, Diez, J, Schulze-Briese, C, Schluckebier, G. | | 登録日 | 2007-12-14 | | 公開日 | 2008-09-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Ultralente--A Microcrystalline Insulin Suspension.
Proteins, 74, 2009
|
|
4CW5
 
 | |
4AK0
 
 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | 分子名称: | INSULIN A CHAIN, INSULIN B CHAIN | | 著者 | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | 登録日 | 2012-02-21 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
4EI8
 
 | |
4AK1
 
 | | Structure of BT4661, a SusE-like surface located polysaccharide binding protein from the Bacteroides thetaiotaomicron heparin utilisation locus | | 分子名称: | BT_4661, SODIUM ION | | 著者 | Lowe, E.C, Basle, A, Czjzek, M, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | 登録日 | 2012-02-21 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4B6F
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | 分子名称: | (4-phenoxyphenyl)methylazanium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | 著者 | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | 登録日 | 2012-08-09 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
4B73
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | 分子名称: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | 著者 | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | 登録日 | 2012-08-16 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
3GX5
 
 | |
2D4V
 
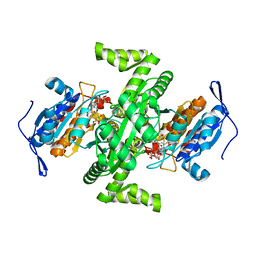 | | Crystal structure of NAD dependent isocitrate dehydrogenase from Acidithiobacillus thiooxidans | | 分子名称: | CITRATE ANION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, isocitrate dehydrogenase | | 著者 | Imada, K, Tamura, T, Namba, K, Inagaki, K. | | 登録日 | 2005-10-24 | | 公開日 | 2006-11-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity
Proteins, 70, 2008
|
|
2XZ2
 
 | |
2XQO
 
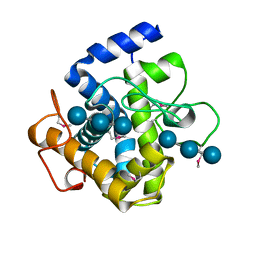 | | CtCel124: a cellulase from Clostridium thermocellum | | 分子名称: | Dockerin type 1, NICKEL (II) ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Carvalho, A.L, Verze, G, Bras, J.L.A, Cartmell, A, Bayer, E.A, Vazana, Y, Correia, M.A.S, Prates, J.A.M, Gilbert, H.J, Fontes, C.M.G.A, Romao, M.J. | | 登録日 | 2010-09-06 | | 公開日 | 2011-03-02 | | 最終更新日 | 2021-02-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Insights Into a Unique Cellulase Fold and Mechanism of Cellulose Hydrolysis
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3H3D
 
 | |
3GX2
 
 | |
3GX6
 
 | |
2EZK
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, REGULARIZED MEAN STRUCTURE | | 分子名称: | TRANSPOSASE | | 著者 | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | 登録日 | 1997-10-04 | | 公開日 | 1998-01-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2EZL
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, 29 STRUCTURES | | 分子名称: | TRANSPOSASE | | 著者 | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | 登録日 | 1997-10-04 | | 公開日 | 1998-01-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
1DKL
 
 | |
1HWG
 
 | |
7YO8
 
 | |
6UDL
 
 | |
