6PX1
 
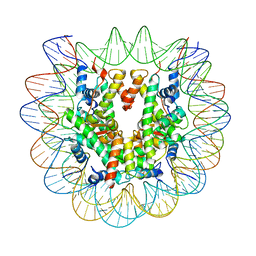 | | Set2 bound to nucleosome | | 分子名称: | DNA (149-MER), Histone H2B 1.1, Histone H3, ... | | 著者 | Halic, M, Bilokapic, S. | | 登録日 | 2019-07-24 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Nucleosome and ubiquitin position Set2 to methylate H3K36.
Nat Commun, 10, 2019
|
|
1LTL
 
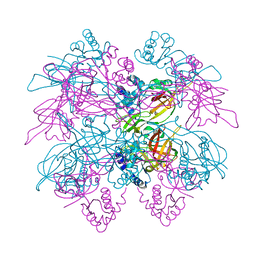 | | THE DODECAMER STRUCTURE OF MCM FROM ARCHAEAL M. THERMOAUTOTROPHICUM | | 分子名称: | DNA replication initiator (Cdc21/Cdc54), ZINC ION | | 著者 | Fletcher, R.J, Bishop, B.E, Leon, R.P, Sclafani, R.A, Ogata, C.M, Chen, X.S. | | 登録日 | 2002-05-20 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Structure and function of MCM from archaeal M. Thermoautotrophicum
Nat.Struct.Biol., 10, 2003
|
|
1ZT2
 
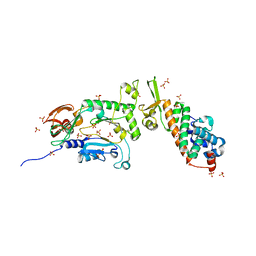 | | Heterodimeric structure of the core primase. | | 分子名称: | DNA primase large subunit, DNA primase small subunit, SULFATE ION, ... | | 著者 | Lao-Sirieix, S.H, Nookala, R.K, Roversi, P, Bell, S.D, Pellegrini, L. | | 登録日 | 2005-05-26 | | 公開日 | 2005-11-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | Structure of the heterodimeric core primase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3KBD
 
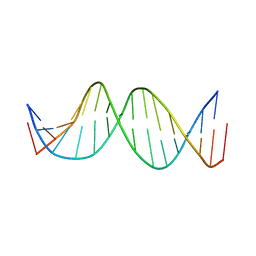 | | MUTATED NF KAPPA-B SITE, BI MODEL | | 分子名称: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*GP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*CP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | 著者 | Tisne, C, Hartmann, B, Delepierre, M. | | 登録日 | 1998-11-30 | | 公開日 | 1999-10-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NF-kappa B binding mechanism: a nuclear magnetic resonance and modeling study of a GGG --> CTC mutation.
Biochemistry, 38, 1999
|
|
1BHR
 
 | | 2'-DEOXY-ISOGUANOSINE BASE PAIRED TO THYMIDINE, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*CP*GP*CP*IGUP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | 著者 | Robinson, H, Gao, Y.-G, Bauer, C, Roberts, C, Switzer, C, Wang, A.H.-J. | | 登録日 | 1998-06-10 | | 公開日 | 1998-11-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 2'-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis.
Biochemistry, 37, 1998
|
|
5ZEV
 
 | |
1NP7
 
 | | Crystal Structure Analysis of Synechocystis sp. PCC6803 cryptochrome | | 分子名称: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Brudler, R, Hitomi, K, Daiyasu, H, Toh, H, Kucho, K, Ishiura, M, Kanehisa, M, Roberts, V.A, Todo, T, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2003-01-17 | | 公開日 | 2003-01-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of a new cryptochrome class: structure, function, and evolution
Mol.Cell, 11, 2003
|
|
8OZD
 
 | | cryoEM structure of SPARTA complex dimer-3 | | 分子名称: | DNA (16-MER), Piwi domain-containing protein, RNA (18-MER), ... | | 著者 | Ekundayo, B, Ni, D.C, Lu, X.H, Stahlberg, H. | | 登録日 | 2023-05-09 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | ELECTRON MICROSCOPY (3.89 Å) | | 主引用文献 | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
2DFL
 
 | | Crystal structure of left-handed RadA filament | | 分子名称: | DNA repair and recombination protein radA | | 著者 | Chen, L.T, Ko, T.P, Wang, T.F, Wang, A.H.J. | | 登録日 | 2006-03-02 | | 公開日 | 2007-01-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the left-handed archaeal RadA helical filament: identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins
Nucleic Acids Res., 35, 2007
|
|
1NHJ
 
 | | Crystal structure of N-terminal 40KD MutL/A100P mutant protein complex with ADPnP and one sodium | | 分子名称: | DNA mismatch repair protein mutL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Hu, X, Machius, M, Yang, W. | | 登録日 | 2002-12-19 | | 公開日 | 2003-06-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
3W03
 
 | | XLF-XRCC4 complex | | 分子名称: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | 著者 | Wu, Q, Ochi, T, Matak-Vinkovic, D, Robinson, C.V, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2012-10-17 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (8.492 Å) | | 主引用文献 | Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions
Biochem.Soc.Trans., 39, 2011
|
|
2RHF
 
 | | D. radiodurans RecQ HRDC domain 3 | | 分子名称: | DNA helicase RecQ, PHOSPHATE ION | | 著者 | Keck, J.L, Killoran, M.P. | | 登録日 | 2007-10-09 | | 公開日 | 2008-04-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ.
Nucleic Acids Res., 36, 2008
|
|
8J92
 
 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana H2A.W | | 分子名称: | DNA (169-MER), HTA6, HTB9, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
2PIS
 
 | | Efforts toward Expansion of the Genetic Alphabet: Structure and Replication of Unnatural Base Pairs | | 分子名称: | DNA (5'-D(*CP*GP*(CBR)P*GP*AP*AP*(FFD)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Matsuda, S, Fillo, J.D, Henry, A.A, Wilkins, S.J, Rai, P, Dwyer, T.J, Geierstanger, B.H, Wemmer, D.E, Schultz, P.G, Spraggon, G, Romesberg, F.E. | | 登録日 | 2007-04-13 | | 公開日 | 2007-10-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs.
J.Am.Chem.Soc., 129, 2007
|
|
8KFT
 
 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 15 seconds | | 分子名称: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFS
 
 | | Crystal structure of ZmMOC1/nicked Holliday junction complex at ground state | | 分子名称: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
3OLC
 
 | |
4KSH
 
 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (7-({4-[(3R)-3-AMINOPYRROLIDIN-1-YL]-5-CHLORO-6-ETHYL-7H-PYRROLO[2,3-D]PYRIMIDIN-2-YL}SULFANYL)-1,5-NAPHTHYRIDIN-1(4H)-OL) | | 分子名称: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)-1,5-naphthyridin-1(4H)-ol, DNA gyrase subunit B, GLYCEROL | | 著者 | Bensen, D.C, Akers-Rodriguez, S, Tari, L.W. | | 登録日 | 2013-05-17 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A new class of type iia topoisomerase inhibitors with broad-spectrum antibacterial activity
To be Published
|
|
2N1G
 
 | |
2N60
 
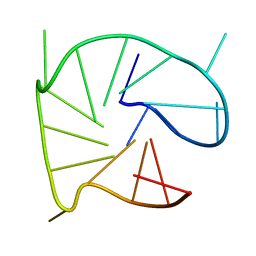 | | G-quadruplexes with (4n-1) guanines in the G-tetrad core: formation of a G-triad water complex and implication for small-molecule binding | | 分子名称: | DNA (5'-D(*TP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*T)-3') | | 著者 | Heddi, B, Martin-Pintado, N, Serimbetov, Z, Kari, T.M, Phan, A.T. | | 登録日 | 2015-08-08 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | G-quadruplexes with (4n - 1) guanines in the G-tetrad core: formation of a G-triadwater complex and implication for small-molecule binding
Nucleic Acids Res., 44, 2016
|
|
2LFY
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dA | | 分子名称: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | 著者 | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | 登録日 | 2011-07-18 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
7DOR
 
 | |
7DPR
 
 | |
7DPS
 
 | |
7DQH
 
 | |
