2XGR
 
 | |
1BXI
 
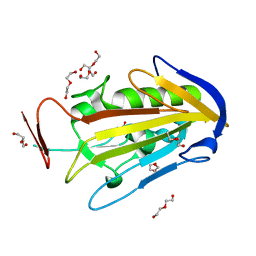
 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 | | 分子名称: | NICKEL (II) ION, PHOSPHATE ION, PROTEIN (COLICIN E9 IMMUNITY PROTEIN), ... | | 著者 | Kuhlmann, U.C, Kleanthous, C, James, R, Moore, G.R, Hemmings, A.M. | | 登録日 | 1998-10-04 | | 公開日 | 1999-10-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal Structure of the E.Coli Colicin E9 DNase Domain With its Cognate Immunity Protein Im9
Thesis, 1998
|
|
4DFI
 
 | | Crystal structure of cell adhesion molecule nectin-2/CD112 mutant FAMP | | 分子名称: | Poliovirus receptor-related protein 2 | | 著者 | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | 登録日 | 2012-01-23 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
4DFH
 
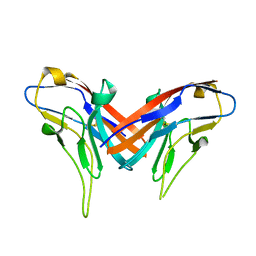 | | Crystal structure of cell adhesion molecule nectin-2/CD112 variable domain | | 分子名称: | Poliovirus receptor-related protein 2 | | 著者 | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | 登録日 | 2012-01-23 | | 公開日 | 2012-06-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
6ISA
 
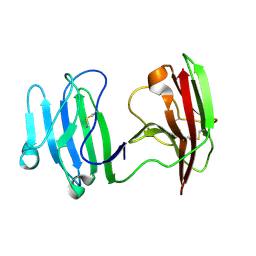 | | mCD226 | | 分子名称: | CD226 antigen | | 著者 | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | 登録日 | 2018-11-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5K4C
 
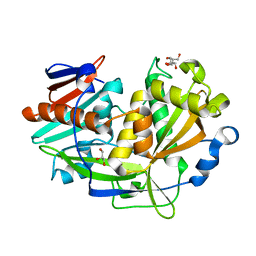 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 2 | | 分子名称: | Eukaryotic translation initiation factor 3 subunit D, GLYCEROL | | 著者 | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | 登録日 | 2016-05-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
6VZD
 
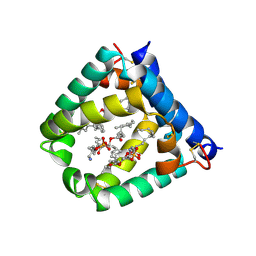 | | N-terminal domain of mouse surfactant protein B (K46E/R51E mutant) with bound lipid | | 分子名称: | (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphapentacos-7-en-19-yl (9Z)-octadec-9-enoate, Pulmonary surfactant-associated protein B | | 著者 | Rapoport, T.A, Bodnar, N.O. | | 登録日 | 2020-02-28 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Mechanism of Lamellar Body Formation by Lung Surfactant Protein B.
Mol.Cell, 81, 2021
|
|
7TJ0
 
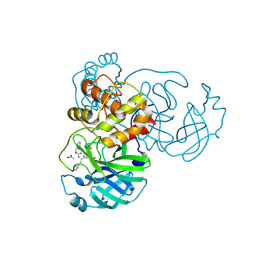 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
6VYN
 
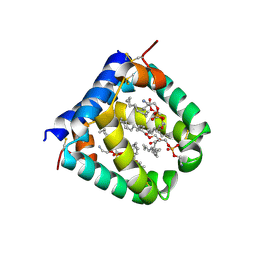 | | N-terminal domain of mouse surfactant protein B with bound lipid, wild type | | 分子名称: | (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphapentacos-7-en-19-yl (9Z)-octadec-9-enoate, Pulmonary surfactant-associated protein B | | 著者 | Rapoport, T.A, Bodnar, N.O. | | 登録日 | 2020-02-27 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-01-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanism of Lamellar Body Formation by Lung Surfactant Protein B.
Mol.Cell, 81, 2021
|
|
6W1B
 
 | |
6VZ0
 
 | |
6VZE
 
 | |
4PR6
 
 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | 分子名称: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | 著者 | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | 登録日 | 2014-03-05 | | 公開日 | 2014-10-29 | | 最終更新日 | 2014-11-26 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
5K4B
 
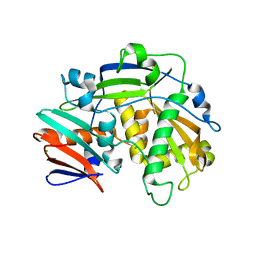 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 1 | | 分子名称: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit D | | 著者 | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | 登録日 | 2016-05-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K4D
 
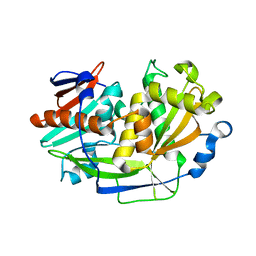 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | 分子名称: | Eukaryotic translation initiation factor 3 subunit D | | 著者 | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | 登録日 | 2016-05-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
7TIV
 
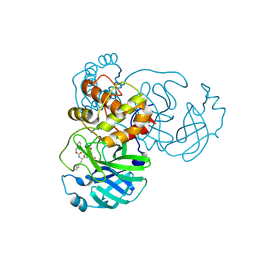 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB48 | | 分子名称: | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, MAGNESIUM ION | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIU
 
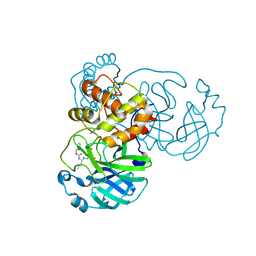 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB46 | | 分子名称: | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, MAGNESIUM ION, ... | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIY
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-48 | | 分子名称: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[(2,4,5-trifluorophenyl)methoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIZ
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-63 | | 分子名称: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[3-(trifluoromethyl)phenyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIW
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB54 | | 分子名称: | (1S,2S)-2-[(N-{[(2-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, PHOSPHATE ION | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIA
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-14 | | 分子名称: | 3C-like proteinase nsp5, THIOCYANATE ION, benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIX
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB56 | | 分子名称: | 3C-like proteinase nsp5, MAGNESIUM ION, N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
4PRF
 
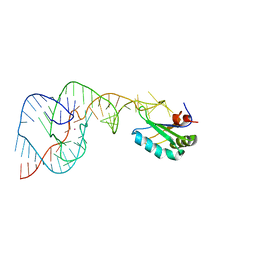 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | 分子名称: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | 著者 | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | 登録日 | 2014-03-05 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.395 Å) | | 主引用文献 | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
2KSW
 
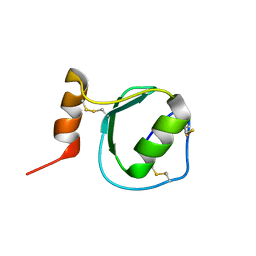 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for Oryctin | | 分子名称: | Oryctin | | 著者 | Horita, S, Ishibashi, J, Nagata, K, Miyakawa, T, Yamakawa, M, Tanokura, M. | | 登録日 | 2010-01-14 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Isolation, cDNA cloning, and structure-based functional characterization of oryctin, a hemolymph protein from the coconut rhinoceros beetle, Oryctes rhinoceros, as a novel serine protease inhibitor
J.Biol.Chem., 285, 2010
|
|
2YUA
 
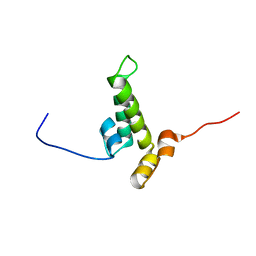 | | Solution structure of the DnaJ domain from human Williams-Beuren syndrome chromosome region 18 protein | | 分子名称: | Williams-Beuren syndrome chromosome region 18 protein | | 著者 | Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-06 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DnaJ domain from human Williams-Beuren syndrome chromosome region 18 protein
To be Published
|
|
