4ZSG
 
 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | 分子名称: | 3-amino-5-[(4-chlorophenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | 著者 | Tucker, J, Ogg, D.J. | | 登録日 | 2015-05-13 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3I17
 
 | |
6JNA
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
1J3G
 
 | | Solution structure of Citrobacter Freundii AmpD | | 分子名称: | AmpD protein, ZINC ION | | 著者 | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | 登録日 | 2003-01-31 | | 公開日 | 2003-02-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
2J5I
 
 | | Crystal Structure of Hydroxycinnamoyl-CoA Hydratase-Lyase | | 分子名称: | P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | 著者 | Leonard, P.M, Brzozowski, A.M, Lebedev, A, Marshall, C.M, Smith, D.J, Verma, C.S, Walton, N.J, Grogan, G. | | 登録日 | 2006-09-18 | | 公開日 | 2006-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The 1.8 A Resolution Structure of Hydroxycinnamoyl- Coenzyme a Hydratase-Lyase (Hchl) from Pseudomonas Fluorescens, an Enzyme that Catalyses the Transformation of Feruloyl-Coenzyme a to Vanillin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1LOK
 
 | | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris: A Tale of Buffer Inhibition | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, SODIUM ION, ... | | 著者 | Desmarais, W.T, Bienvenue, D.L, Bzymek, K.P, Holz, R.C, Petsko, G.A, Ringe, D. | | 登録日 | 2002-05-06 | | 公開日 | 2002-11-27 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris A tale of Buffer Inhibition
Structure, 10, 2002
|
|
6JN9
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JNC
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
2PDM
 
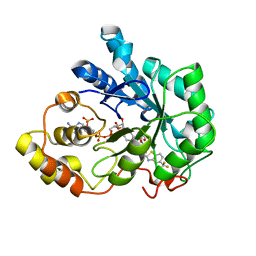 | | Human aldose reductase mutant S302R complexed with zopolrestat. | | 分子名称: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Steuber, H, Heine, A, Klebe, G. | | 登録日 | 2007-04-01 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
4PR6
 
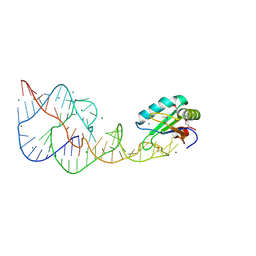 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | 分子名称: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | 著者 | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | 登録日 | 2014-03-05 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
3LQS
 
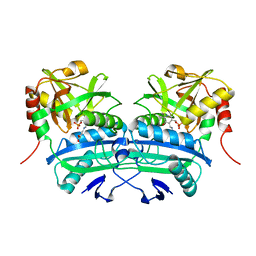 | | Complex Structure of D-Amino Acid Aminotransferase and 4-amino-4,5-dihydro-thiophenecarboxylic acid (ADTA) | | 分子名称: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, ACETIC ACID, D-alanine aminotransferase | | 著者 | Lepore, B.W, Liu, D, Peng, Y, Fu, M, Yasuda, C, Manning, J.M, Silverman, R.B, Ringe, D. | | 登録日 | 2010-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Chiral discrimination among aminotransferases: inactivation by 4-amino-4,5-dihydrothiophenecarboxylic acid.
Biochemistry, 49, 2010
|
|
4BIM
 
 | |
1R48
 
 | | Solution structure of the C-terminal cytoplasmic domain residues 468-497 of Escherichia coli protein ProP | | 分子名称: | Proline/betaine transporter | | 著者 | Zoetewey, D.L, Tripet, B.P, Kutateladze, T.G, Overduin, M.J, Wood, J.M, Hodges, R.S. | | 登録日 | 2003-10-03 | | 公開日 | 2003-12-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the C-terminal Antiparallel Coiled-coil Domain from Escherichia coli Osmosensor ProP.
J.Mol.Biol., 334, 2003
|
|
6JND
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
1XGE
 
 | | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between subunits | | 分子名称: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | 著者 | Lee, M, Chan, C.W, Guss, J.M, Christopherson, R.I, Maher, M.J. | | 登録日 | 2004-09-17 | | 公開日 | 2005-04-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between Subunits
J.Mol.Biol., 348, 2005
|
|
1QI9
 
 | | X-RAY SIRAS STRUCTURE DETERMINATION OF A VANADIUM-DEPENDENT HALOPEROXIDASE FROM ASCOPHYLLUM NODOSUM AT 2.0 A RESOLUTION | | 分子名称: | VANADATE ION, Vanadium-dependent bromoperoxidase | | 著者 | Weyand, M, Hecht, H.-J, Kiess, M, Liaud, M.F, Vilter, H, Schomburg, D. | | 登録日 | 1999-06-10 | | 公開日 | 2000-06-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | X-ray structure determination of a vanadium-dependent haloperoxidase from Ascophyllum nodosum at 2.0 A resolution.
J.Mol.Biol., 293, 1999
|
|
1OE1
 
 | |
3HPA
 
 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | 分子名称: | AMIDOHYDROLASE, ZINC ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-06-03 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
3I40
 
 | | Human insulin | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Timofeev, V.I, Bezuglov, V.V, Miroshnikov, K.A, Chuprov-Netochin, R.N, Kuranova, I.P. | | 登録日 | 2009-07-01 | | 公開日 | 2010-01-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | X-ray investigation of gene-engineered human insulin crystallized from a solution containing polysialic acid.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3MPC
 
 | | The crystal structure of a Fn3-like protein from Clostridium thermocellum | | 分子名称: | Fn3-like protein, SULFATE ION | | 著者 | Alahuhta, M.P, Xu, Q, Brunecky, R, Lunin, V.V. | | 登録日 | 2010-04-26 | | 公開日 | 2010-08-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of a fibronectin type III-like module from Clostridium thermocellum.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5OQ1
 
 | | Crystal structure of Serratia marcescens ChiX (used as MR model for superior PDB 5OPZ) | | 分子名称: | CHLORIDE ION, ChiX, ZINC ION | | 著者 | Owen, R.A, Fyfe, P.K, Lodge, A, Biboy, J, Vollmer, W, Hunter, W.N, Sargent, F. | | 登録日 | 2017-08-10 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structure and activity of ChiX: a peptidoglycan hydrolase required for chitinase secretion by Serratia marcescens.
Biochem. J., 475, 2018
|
|
4ZSJ
 
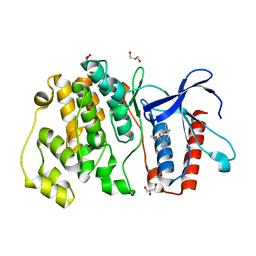 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | 分子名称: | 3-amino-5-[(4-chloro-3-methylphenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | 著者 | Tucker, J, Ogg, D.J. | | 登録日 | 2015-05-13 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3LQE
 
 | |
1I8J
 
 | | CRYSTAL STRUCTURE OF PORPHOBILINOGEN SYNTHASE COMPLEXED WITH THE INHIBITOR 4,7-DIOXOSEBACIC ACID | | 分子名称: | 4,7-DIOXOSEBACIC ACID, MAGNESIUM ION, PORPHOBILINOGEN SYNTHASE, ... | | 著者 | Kervinen, J, Jaffe, E.K, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | 登録日 | 2001-03-14 | | 公開日 | 2001-06-20 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mechanistic basis for suicide inactivation of porphobilinogen synthase by 4,7-dioxosebacic acid, an inhibitor that shows dramatic species selectivity.
Biochemistry, 40, 2001
|
|
5BYZ
 
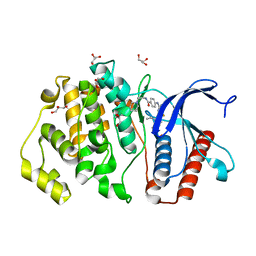 | | ERK5 in complex with small molecule | | 分子名称: | 4-({5-fluoro-4-[2-methyl-1-(propan-2-yl)-1H-imidazol-5-yl]pyrimidin-2-yl}amino)-N-[2-(piperidin-1-yl)ethyl]benzamide, GLYCEROL, Mitogen-activated protein kinase 7 | | 著者 | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | 登録日 | 2015-06-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
