2GEY
 
 | | Crystal Structure of AclR a putative hydroxylase from Streptomyces galilaeus | | 分子名称: | AclR protein, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Beinker, P, Lohkamp, B, Schneider, G. | | 登録日 | 2006-03-21 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of SnoaL2 and AclR: two putative hydroxylases in the biosynthesis of aromatic polyketide antibiotics
J.Mol.Biol., 359, 2006
|
|
5I63
 
 | |
5I01
 
 | | Structure of phosphoheptose isomerase GmhA from Neisseria gonorrhoeae | | 分子名称: | Phosphoheptose isomerase, ZINC ION | | 著者 | Wierzbicki, I.H, Sikora, A.E, Korotkov, K.V. | | 登録日 | 2016-02-03 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Functional and structural studies on the Neisseria gonorrhoeae GmhA, the first enzyme in the glycero-manno-heptose biosynthesis pathways, demonstrate a critical role in lipooligosaccharide synthesis and gonococcal viability.
Microbiologyopen, 6, 2017
|
|
2G95
 
 | |
5I6P
 
 | |
2G9T
 
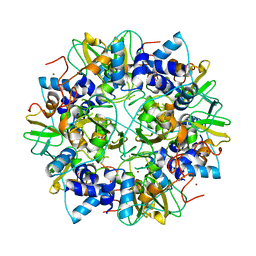 | | Crystal structure of the SARS coronavirus nsp10 at 2.1A | | 分子名称: | ZINC ION, orf1a polyprotein | | 著者 | Su, D, Lou, Z, Yang, H, Sun, F, Rao, Z. | | 登録日 | 2006-03-07 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
2GGT
 
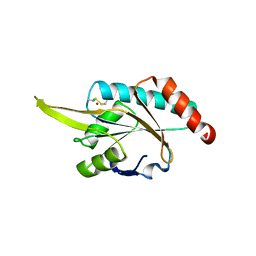 | | Crystal structure of human SCO1 complexed with nickel. | | 分子名称: | CHLORIDE ION, NICKEL (II) ION, SCO1 protein homolog, ... | | 著者 | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Martinelli, M, Palumaa, P, Wang, S. | | 登録日 | 2006-03-24 | | 公開日 | 2006-05-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7D32
 
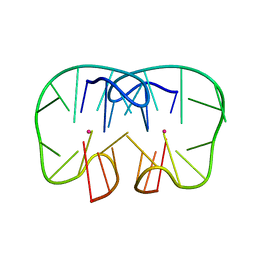 | | The TBA-Pb2+ complex in P41212 space group | | 分子名称: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | 著者 | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | 登録日 | 2020-09-18 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.707 Å) | | 主引用文献 | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
2GHG
 
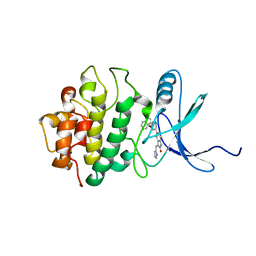 | | h-CHK1 complexed with A431994 | | 分子名称: | 5-{5-[(S)-2-AMINO-3-(1H-INDOL-3-YL)-PROPOXYL]-PYRIDIN-3-YL}-3-[1-(1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Serine/threonine-protein kinase Chk1 | | 著者 | Park, C. | | 登録日 | 2006-03-27 | | 公開日 | 2007-03-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Discovery and SAR of oxindole-pyridine-based protein kinase B/Akt inhibitors for treating cancers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2RUH
 
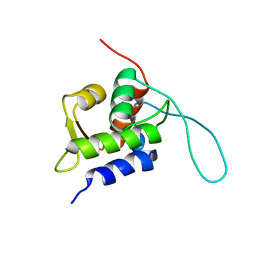 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | 分子名称: | E3 ubiquitin-protein ligase Mdm2 | | 著者 | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | 登録日 | 2014-06-03 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
2RSO
 
 | |
5I1V
 
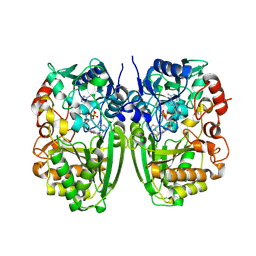 | |
2ROT
 
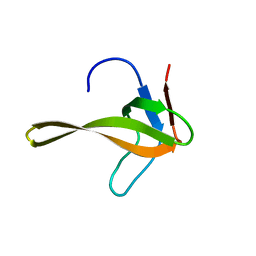 | | Structure of chimeric variant of SH3 domain- SHH | | 分子名称: | Spectrin alpha chain, brain | | 著者 | Kutyshenko, N.P, Prokhorov, D.A, Timchenko, M.A, Kudrevatykh, Y.A, Gushchina, L.V, Khristoforov, V.S, Filimonov, V.V. | | 登録日 | 2008-04-10 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the chimeric SH3 domains, SHH- and SHA-"Bergeracs".
Biochim.Biophys.Acta, 1794, 2009
|
|
7D31
 
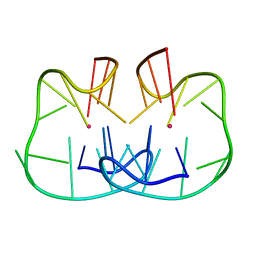 | | The TBA-Pb2+ complex in P41212 space group | | 分子名称: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | 著者 | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | 登録日 | 2020-09-18 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.396 Å) | | 主引用文献 | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
2GHZ
 
 | |
2RUP
 
 | | Solution structure of rat P2X4 receptor head domain | | 分子名称: | P2X purinoceptor 4 | | 著者 | Abe, Y, Igawa, T, Tsuda, M, Inoue, K, Ueda, T. | | 登録日 | 2014-11-12 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the rat P2X4 receptor head domain involved in inhibitory metal binding
FEBS Lett., 589, 2015
|
|
2GGR
 
 | |
5I3V
 
 | |
5I92
 
 | |
2GHE
 
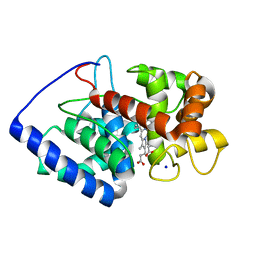 | | Conformational mobility in the active site of a heme peroxidase | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, cytosolic ascorbate peroxidase 1 | | 著者 | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | 登録日 | 2006-03-27 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
7D33
 
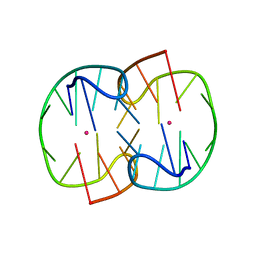 | | The Pb2+ complexed structure of TBA G8C mutant | | 分子名称: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*CP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | 著者 | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | 登録日 | 2020-09-18 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.117 Å) | | 主引用文献 | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
5I5K
 
 | |
2REK
 
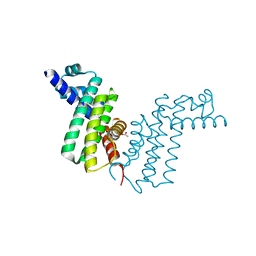 | | Crystal structure of tetR-family transcriptional regulator | | 分子名称: | ACETATE ION, Putative tetR-family transcriptional regulator | | 著者 | Dong, A, Xu, X, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-09-26 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of tetR-family transcriptional regulator.
To be Published
|
|
5I7W
 
 | |
2RER
 
 | |
