8HVC
 
 | |
8HVD
 
 | |
5KQX
 
 | | Protease E35D-SQV | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease E35D-SQV | | 著者 | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | 登録日 | 2016-07-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KR5
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-07 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
8FYO
 
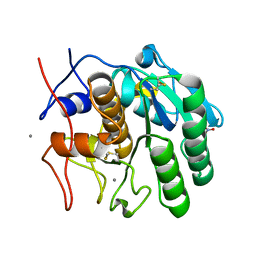 | | MicroED structure of Proteinase K from lamellae milled from multiple plasma sources | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | 登録日 | 2023-01-26 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.39 Å) | | 主引用文献 | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
4YIC
 
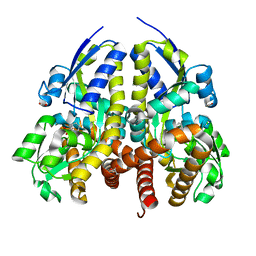 | | CRYSTAL STRUCTURE OF A TRAP TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BORDETELLA BRONCHISEPTICA RB50 (BB0280, TARGET EFI-500035) WITH BOUND PICOLINIC ACID | | 分子名称: | ACETATE ION, CALCIUM ION, IMIDAZOLE, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2015-03-01 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF A TRAP TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BORDETELLA BRONCHISEPTICA RB50 (BB0280, TARGET EFI-500035) WITH BOUND PICOLINIC ACID
To be published
|
|
8FHV
 
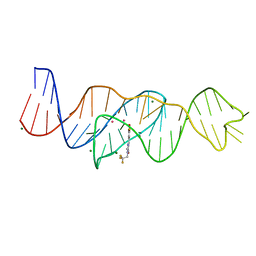 | | Structure of Lettuce aptamer bound to DFHBI-1T with thallium I ions | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
5KT1
 
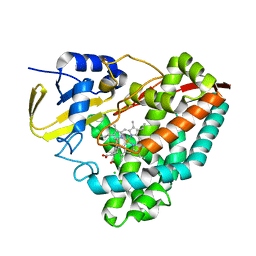 | |
8HUG
 
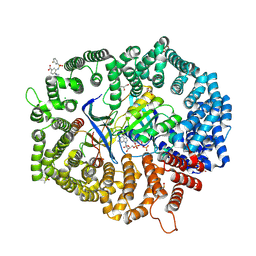 | | F1 in complex with CRM1-Ran-RanBP1 | | 分子名称: | 4-[4-(3-chlorophenyl)piperazin-1-yl]-3-[(3-fluorophenyl)sulfonylamino]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | 著者 | Sun, Q, Lei, Y. | | 登録日 | 2022-12-23 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
5KV0
 
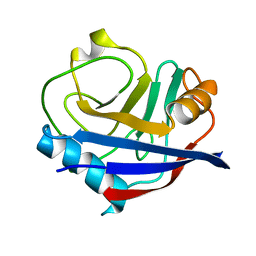 | | Human cyclophilin A at 278K, Data set 2 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | 登録日 | 2016-07-13 | | 公開日 | 2016-08-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
4Y9I
 
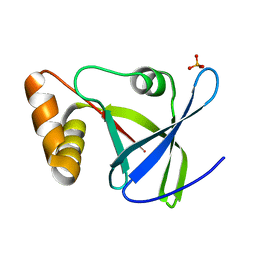 | |
5KV9
 
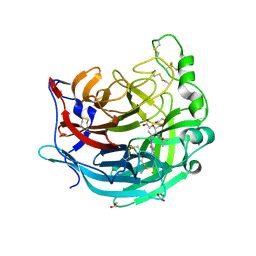 | | Crystal structure of a hPIV haemagglutinin-neuraminidase-inhibitor complex | | 分子名称: | 1,2-ETHANEDIOL, 2,6-anhydro-3,4,5-trideoxy-5-[(2-methylpropanoyl)amino]-4-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-D-galacto-non-2-en onic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Dirr, L, El-Deeb, I.M, Chavas, L.M.G, Guillon, P, von Itzstein, M. | | 登録日 | 2016-07-13 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The impact of the butterfly effect on human parainfluenza virus haemagglutinin-neuraminidase inhibitor design.
Sci Rep, 7, 2017
|
|
8FXF
 
 | | Crystal structure of the coiled-coil domain of TRIM56 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase TRIM56 | | 著者 | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | 登録日 | 2023-01-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | TRIM56 coiled-coil domain structure provides insights into its E3 ligase functions.
Comput Struct Biotechnol J, 21, 2023
|
|
4YMK
 
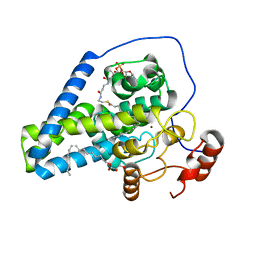 | | Crystal Structure of Stearoyl-Coenzyme A Desaturase 1 | | 分子名称: | Acyl-CoA desaturase 1, STEAROYL-COENZYME A, ZINC ION, ... | | 著者 | Bai, Y, McCoy, J.G, Rajashankar, K.R, Zhou, M. | | 登録日 | 2015-03-06 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.605 Å) | | 主引用文献 | X-ray structure of a mammalian stearoyl-CoA desaturase.
Nature, 524, 2015
|
|
6W3C
 
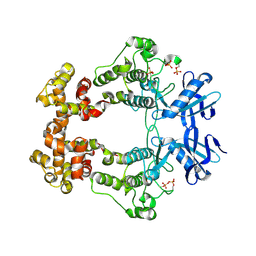 | | Structure of phosphorylated apo IRE1 | | 分子名称: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | 著者 | Wallweber, H, Mortara, K, Ferri, E, Wang, W, Rudolph, J. | | 登録日 | 2020-03-09 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
6W3X
 
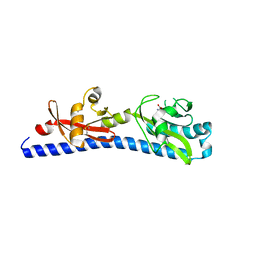 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-valine | | 分子名称: | GLYCEROL, Methyl-accepting chemotaxis protein, SULFATE ION, ... | | 著者 | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
8F93
 
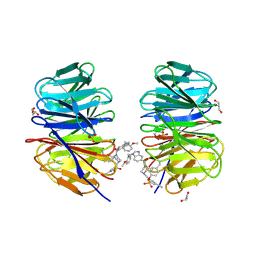 | | WDR5 covalently modified at Y228 by (R)-2-SF | | 分子名称: | 3-ethynyl-5-{[(3R)-4-{1-[(2-methoxyphenyl)methyl]-1H-benzimidazole-5-carbonyl}-3-methylpiperazin-1-yl]methyl}benzene-1-sulfonyl fluoride, CHLORIDE ION, GLYCEROL, ... | | 著者 | Taunton, J, Craven, G.B, Chen, Y. | | 登録日 | 2022-11-23 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Direct mapping of ligandable tyrosines and lysines in cells with chiral sulfonyl fluoride probes.
Nat.Chem., 15, 2023
|
|
6VYJ
 
 | | Human UHRF1 TTD domain in complex with a fragment | | 分子名称: | 2,4-dimethylpyridine, E3 ubiquitin-protein ligase UHRF1, beta-D-glucopyranose | | 著者 | Campbell, J.C, Chang, L, Young, D.W. | | 登録日 | 2020-02-26 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Discovery of small molecules targeting the tandem tudor domain of the epigenetic factor UHRF1 using fragment-based ligand discovery.
Sci Rep, 11, 2021
|
|
5KZW
 
 | | Crystal structure of human GAA | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Deming, D.T, Garman, S.C. | | 登録日 | 2016-07-25 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of human GAA: structural basis of Pompe disease
To be published
|
|
8FI2
 
 | | Structure of Lettuce C20T bound to DFHBI-1T | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
4YCP
 
 | | E. coli dihydrouridine synthase C (DusC) in complex with tRNATrp | | 分子名称: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Byrne, R.T, Jenkins, H.T, Peters, D.T, Whelan, F, Stowell, J, Aziz, N, Kasatsky, P, Rodnina, M.V, Koonin, E.V, Konevega, A.L, Antson, A.A. | | 登録日 | 2015-02-20 | | 公開日 | 2015-04-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8HU4
 
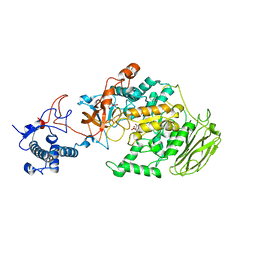 | | Limosilactobacillus reuteri N1 GtfB | | 分子名称: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Dong, J.J, Bai, Y.X. | | 登録日 | 2022-12-22 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
6WAG
 
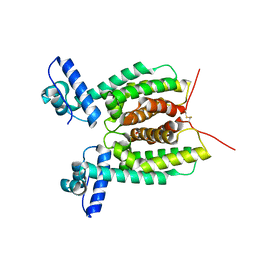 | | Crystal Structure of SmcR S76A from Vibrio Vulnificus | | 分子名称: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | 著者 | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | 登録日 | 2020-03-25 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.575 Å) | | 主引用文献 | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
5KYB
 
 | |
6WAU
 
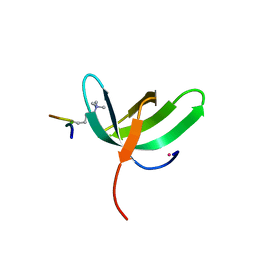 | | Complex structure of PHF19 | | 分子名称: | Histone H3.1t peptide, PHD finger protein 19, UNKNOWN ATOM OR ION | | 著者 | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | 登録日 | 2020-03-26 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
