8FN5
 
 | |
5LBO
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-001 | | 分子名称: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | 著者 | Singh, A.K, Brown, D.G. | | 登録日 | 2016-06-16 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
6W5L
 
 | | 2.1 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7g | | 分子名称: | (2~{S})-~{N}-[(1~{R})-1-[bis($l^{1}-oxidanyl)-methoxy-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[[2-(3-chlorophenyl)-2-methyl-propoxy]-oxidanylidene-methyl]amino]-4-methyl-pentanamide, 3C-LIKE PROTEASE | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6W66
 
 | | The structure of the F64A, S172A mutant Keap1-BTB domain in complex with SKP1-FBXL17 | | 分子名称: | F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, S-phase kinase-associated protein 1 | | 著者 | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | 登録日 | 2020-03-16 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
4YR1
 
 | |
4YRI
 
 | |
4YRR
 
 | |
8FN3
 
 | |
6W83
 
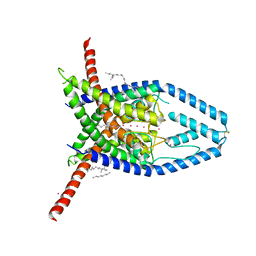 | | K2P2.1 (TREK-1), 100 mM K+ | | 分子名称: | CADMIUM ION, DECANE, DODECANE, ... | | 著者 | Lolicato, M, Minor, D.L. | | 登録日 | 2020-03-20 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.901 Å) | | 主引用文献 | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
8FI8
 
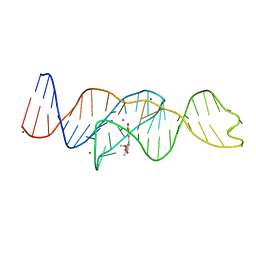 | |
6W87
 
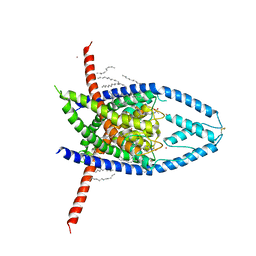 | |
5KKK
 
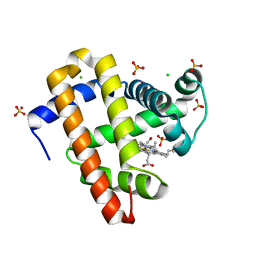 | |
6W8R
 
 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | 分子名称: | Metacaspase-4, SULFATE ION | | 著者 | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | 登録日 | 2020-03-21 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
8HT0
 
 | |
6W9I
 
 | |
5KM6
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant Ara-A nucleoside phosphoramidate substrate complex | | 分子名称: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[2-(1~{H}-indol-3-yl)ethyl]phosphonamidic acid | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-06-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KM2
 
 | |
4YWF
 
 | | AbyA5 | | 分子名称: | AbyA5 | | 著者 | Byrne, M.J, Race, P.R. | | 登録日 | 2015-03-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2019-06-19 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An Esterase-like Lyase Catalyzes Acetate Elimination in Spirotetronate/Spirotetramate Biosynthesis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8FO6
 
 | | Nucleotide-free structure of a functional construct of eukaryotic elongation factor 2 kinase. | | 分子名称: | CALCIUM ION, Calmodulin-1, Eukaryotic elongation factor 2 kinase, ... | | 著者 | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | 登録日 | 2022-12-29 | | 公開日 | 2023-05-03 | | 実験手法 | X-RAY DIFFRACTION (2.553 Å) | | 主引用文献 | ADP enhances the allosteric activation of eukaryotic elongation factor 2 kinase by calmodulin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5KMR
 
 | |
5KNJ
 
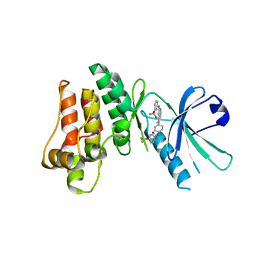 | | Pseudokinase Domain of MLKL bound to Compound 1. | | 分子名称: | 1-[4-[methyl-[2-[(3-sulfamoylphenyl)amino]pyrimidin-4-yl]amino]phenyl]-3-[4-(trifluoromethyloxy)phenyl]urea, Mixed lineage kinase domain-like protein | | 著者 | Marcotte, D.J. | | 登録日 | 2016-06-28 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | ATP-Competitive MLKL Binders Have No Functional Impact on Necroptosis.
Plos One, 11, 2016
|
|
8HUH
 
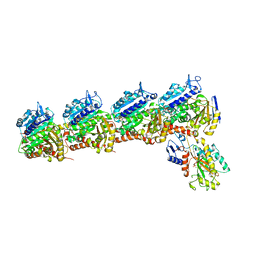 | | Crystal structure of T2R-TTL-3a complex | | 分子名称: | 2-(1-methylindol-4-yl)-4-(3,4,5-trimethoxyphenyl)-1~{H}-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y.X, Chen, J.J. | | 登録日 | 2022-12-23 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of T2R-TTL-3a complex
To Be Published
|
|
8FHZ
 
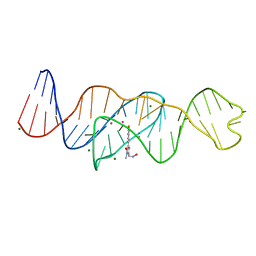 | | Structure of Lettuce aptamer bound to DFHO | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8HVB
 
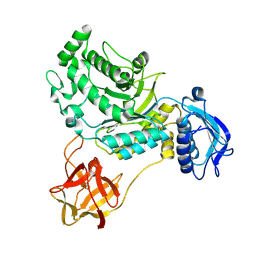 | |
5KOP
 
 | | Arabidopsis thaliana fucosyltransferase 1 (FUT1) in its apo-form | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Galactoside 2-alpha-L-fucosyltransferase | | 著者 | Rocha, J, de Sanctis, D, Breton, C. | | 登録日 | 2016-07-01 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-10-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation.
Plant Cell, 28, 2016
|
|
