5Y1Q
 
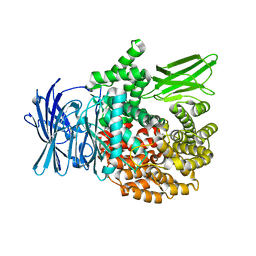 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | 分子名称: | (2S)-2-[(3-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, M1 family aminopeptidase, MAGNESIUM ION, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-21 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5IJW
 
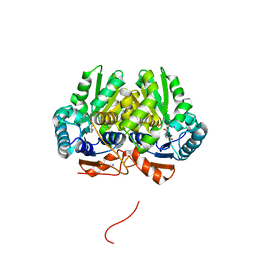 | | Glutamate Racemase (MurI) from Mycobacterium smegmatis with bound D-glutamate, 1.8 Angstrom resolution, X-ray diffraction | | 分子名称: | D-GLUTAMIC ACID, Glutamate racemase, IODIDE ION | | 著者 | Poen, S, Nakatani, Y, Krause, K. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Exploring the structure of glutamate racemase from Mycobacterium tuberculosis as a template for anti-mycobacterial drug discovery.
Biochem. J., 473, 2016
|
|
7T70
 
 | |
7T9Y
 
 | |
7TA7
 
 | |
7T8R
 
 | |
5Y2E
 
 | |
7TB2
 
 | |
7TBT
 
 | |
7TC4
 
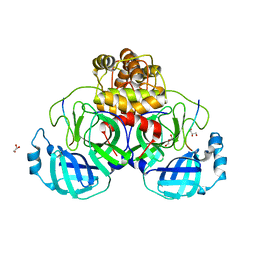 | |
5YGY
 
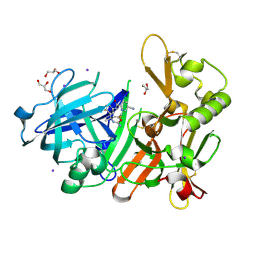 | | Crystal Structure of BACE1 in complex with (S)-N-(3-(2-amino-6-(fluoromethyl)-4 -methyl-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-cyanopicolinamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Fuchino, K, Mitsuoka, Y, Masui, M, Kurose, N, Yoshida, S, Komano, K, Yamamoto, T, Ogawa, M, Unemura, C, Hosono, M, Ito, H, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Miyajima, H, Hiroyama, S, Koyabu, K, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | 登録日 | 2017-09-27 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Rational Design of Novel 1,3-Oxazine Based beta-Secretase (BACE1) Inhibitors: Incorporation of a Double Bond To Reduce P-gp Efflux Leading to Robust A beta Reduction in the Brain
J. Med. Chem., 61, 2018
|
|
5YIA
 
 | | Crystal Structure of KNI-10343 bound Plasmepsin II (PMII) from Plasmodium falciparum | | 分子名称: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-hydroxyphenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | 著者 | Rathore, I, Mishra, V, Bhaumik, P. | | 登録日 | 2017-10-03 | | 公開日 | 2018-07-11 | | 最終更新日 | 2019-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
7T8M
 
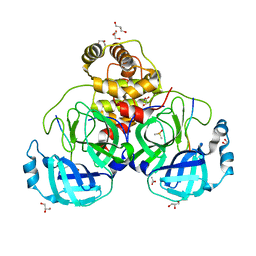 | |
5ILP
 
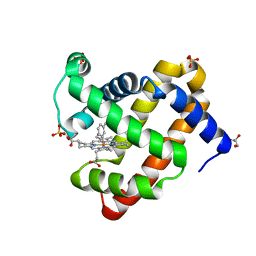 | | H64Q sperm whale myoglobin with a Fe-tolyl moiety | | 分子名称: | GLYCEROL, Myoglobin, SULFATE ION, ... | | 著者 | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | 登録日 | 2016-03-04 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
J. Inorg. Biochem., 164, 2016
|
|
5YJP
 
 | | Human chymase in complex with 3-(ethoxyimino)-7-oxo-1,4-diazepane derivative | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-((R)-1-((R,Z)-6-(5-chloro-2-methoxybenzyl)-3-(ethoxyimino)-7-oxo-1,4-diazepane-1-carboxamido)propyl)benzoic acid, ZINC ION, ... | | 著者 | Sugawara, H. | | 登録日 | 2017-10-11 | | 公開日 | 2017-12-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based design, synthesis, and binding mode analysis of novel and potent chymase inhibitors
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Guo, S, Phippen, S, Campbell, R, Davies, P. | | 登録日 | 2016-03-12 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
7SUV
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite A | | 分子名称: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | 著者 | Whitaker, A.W, Freudenthal, B.D. | | 登録日 | 2021-11-18 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
5IE3
 
 | | Crystal structure of a plant enzyme | | 分子名称: | ADENOSINE MONOPHOSPHATE, OXALIC ACID, Oxalate--CoA ligase | | 著者 | Fan, M.R, Li, M, Chang, W.R. | | 登録日 | 2016-02-24 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
5IEP
 
 | |
5WN5
 
 | |
7SVB
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | 分子名称: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | 著者 | Whitaker, A.W, Freudenthal, B.D. | | 登録日 | 2021-11-18 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
5IOK
 
 | |
5IR3
 
 | |
7L7P
 
 | |
7L7O
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR04-49 | | 分子名称: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2020-12-29 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
