2QHZ
 
 | | Crystal structure of protease inhibitor, MIT-1-AC87 in complex with wild type HIV-1 protease | | 分子名称: | (2E)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-4,4,4-TRIFLUORO-3-METHYLBUT-2-ENAMIDE, PHOSPHATE ION, Protease | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2007-07-03 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QHY
 
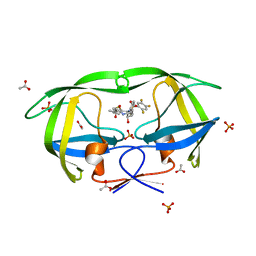 | | Crystal Structure of protease inhibitor, MIT-1-AC86 in complex with wild type HIV-1 protease | | 分子名称: | ACETATE ION, N~2~-ACETYL-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-L-ALANINAMIDE, PHOSPHATE ION, ... | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2007-07-03 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QI0
 
 | |
5EM1
 
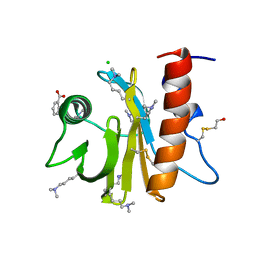 | | Crystal structure of ragweed allergen Amb a 8 | | 分子名称: | BENZOIC ACID, CHLORIDE ION, Profilin | | 著者 | Offermann, L.R, He, J.Z, Perdue, M.L, Chruszcz, M. | | 登録日 | 2015-11-05 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
5EVE
 
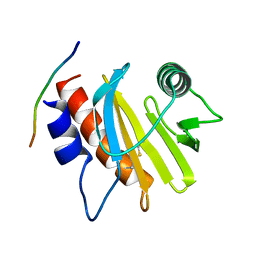 | | Crystal structure of Amb a 8 in complex with poly-Pro10 | | 分子名称: | Poly-Proline peptide, Profilin | | 著者 | Offermann, L.R, Schlachter, C.R, Garrett, J, Chruszcz, M. | | 登録日 | 2015-11-19 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
2MM0
 
 | | Solution Structures of active Ptr ToxB and its inactive homolog highlight protein dynamics as a modulator of toxin activity | | 分子名称: | Host-selective toxin protein | | 著者 | Nyarko, A, Singarapu, K, Figueroa, M, Manning, V, Pandelova, I, Wolpert, T, Ciuffetti, L, Barbar, E. | | 登録日 | 2014-03-06 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structures of Pyrenophora tritici-repentis ToxB and Its Inactive Homolog Reveal Potential Determinants of Toxin Activity.
J.Biol.Chem., 289, 2014
|
|
5FUA
 
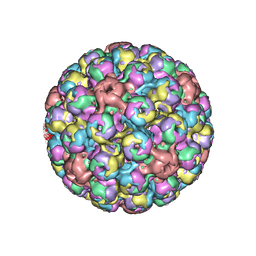 | | Cryo-EM of BK polyomavirus | | 分子名称: | MAJOR CAPSID PROTEIN VP1 | | 著者 | Hurdiss, D.L, Morgan, E.L, Thompson, R.F, Prescott, E.L, Panou, M.M, Macdonald, A, Ranson, N.A. | | 登録日 | 2016-01-22 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | New Structural Insights Into the Genome and Minor Capsid Proteins of Bk Polyomavirus Using Cryo-Electron Microscopy.
Structure, 24, 2016
|
|
5FRR
 
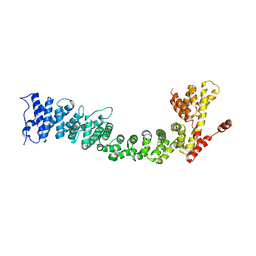 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | 分子名称: | SISTER CHROMATID COHESION PROTEIN PDS5 | | 著者 | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | 登録日 | 2015-12-22 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (5.8 Å) | | 主引用文献 | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
3WHW
 
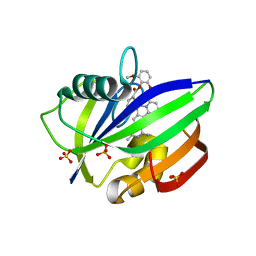 | | MTH1 in complex with Ruthenium-based inhibitor | | 分子名称: | 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION, [4-amino-2-methyl-6-(pyridin-2-yl-kappaN)quinazolin-7-yl-kappaC~7~](carbonyl){1-[(2,6-dimethoxyphenoxy)carbonyl]cyclopenta-2,4-dien-1-yl}ruthenium | | 著者 | Streib, M, Kraeling, K, Richter, K, Steuber, H, Meggers, E. | | 登録日 | 2013-09-03 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | An Organometallic Inhibitor for the Human Repair Enzyme 7,8-Dihydro-8-oxoguanosine Triphosphatase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5G2E
 
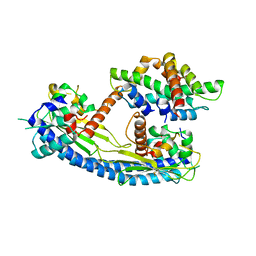 | | Structure of the Nap1 H2A H2B complex | | 分子名称: | HISTONE H2A TYPE 1, HISTONE H2B 1.1, NUCLEOSOME ASSEMBLY PROTEIN | | 著者 | AguilarGurrieri, C, Larabi, A, Vinayachandran, V, Patel, N.A, Yen, K, Reja, R, Ebong, I.O, Schoehn, G, Robinson, C.V, Pugh, B.F, Panne, D. | | 登録日 | 2016-04-07 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (6.7 Å) | | 主引用文献 | Structural Evidence for Nap1-Dependent H2A-H2B Deposition and Nucleosome Assembly.
Embo J., 35, 2016
|
|
5G1S
 
 | | Open conformation of Francisella tularensis ClpP at 1.7 A | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | 著者 | Diaz-Saez, L, Pankov, G, Hunter, W.N. | | 登録日 | 2016-03-30 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Open and compressed conformations of Francisella tularensis ClpP.
Proteins, 85, 2017
|
|
5GMU
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Chlorogenic acid | | 分子名称: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, Protein AroA(G), SULFATE ION | | 著者 | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | 登録日 | 2016-07-16 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
2MUQ
 
 | |
2MZC
 
 | | Metal Binding of Glutaredoxins | | 分子名称: | Glutaredoxin, SILVER ION | | 著者 | Bilinovich, S.M, Caporoso, J.A, Taraboletti, A, Duangjumpa, N, Panzner, M.J, Prokop, J.W, Shriver, L.P, Leeper, T.C. | | 登録日 | 2015-02-11 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Metal Binding of Glutaredoxins
To be Published
|
|
5G64
 
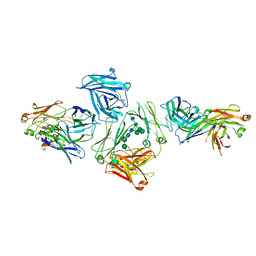 | | The complex between human IgE-Fc and two anti-IgE Fab fragments | | 分子名称: | FAB FRAGMENT, IG EPSILON CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Davies, A.M, Allan, E.G, Keeble, A.H, Delgado, J, Cossins, B.P, Mitropoulou, A.N, Pang, M.O.Y, Ceska, T, Beavil, A.J, Craggs, G, Westwood, M, Henry, A.J, McDonnell, J.M, Sutton, B.J. | | 登録日 | 2016-06-14 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.715 Å) | | 主引用文献 | Allosteric mechanism of action of the therapeutic anti-IgE antibody omalizumab.
J. Biol. Chem., 292, 2017
|
|
5NYW
 
 | | Anbu (ancestral beta-subunit) from Yersinia bercovieri | | 分子名称: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | 著者 | Piasecka, A, Czapinska, H, Vielberg, M, Szczepanowski, R.H, Reed, S, Groll, M, Bochtler, M. | | 登録日 | 2017-05-12 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | The Y. bercovieri Anbu crystal structure sheds light on the evolution of highly (pseudo)symmetric multimers.
J. Mol. Biol., 430, 2018
|
|
2MUR
 
 | |
4NCI
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation | | 分子名称: | DNA double-strand break repair Rad50 ATPase | | 著者 | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | 登録日 | 2013-10-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCJ
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation with ADP Beryllium Flouride | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA double-strand break repair Rad50 ATPase, ... | | 著者 | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | 登録日 | 2013-10-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4DZY
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with ADP | | 分子名称: | (S)-2-chloro-3-phenylpropanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2012-03-01 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2OAA
 
 | | Restriction endonuclease MvaI-cognate DNA substrate complex | | 分子名称: | 5'-D(*CP*AP*TP*CP*CP*AP*GP*GP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*CP*CP*TP*GP*GP*AP*T)-3', CALCIUM ION, ... | | 著者 | Kaus-Drobek, M, Czapinska, H, Sokolowska, M, Tamulaitis, G, Szczepanowski, R.H, Urbanke, K, Siksnys, V, Bochtler, M. | | 登録日 | 2006-12-15 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically.
Nucleic Acids Res., 35, 2007
|
|
5FIS
 
 | | Exonuclease domain-containing 1 (Exd1) in the Gd bound conformation | | 分子名称: | EXD1, GADOLINIUM ATOM | | 著者 | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | 登録日 | 2015-10-02 | | 公開日 | 2015-12-23 | | 最終更新日 | 2016-01-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
4NCK
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R797G mutation | | 分子名称: | CHLORIDE ION, DNA double-strand break repair Rad50 ATPase, MAGNESIUM ION, ... | | 著者 | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | 登録日 | 2013-10-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCH
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 L802W mutation | | 分子名称: | DNA double-strand break repair Rad50 ATPase, SULFATE ION | | 著者 | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | 登録日 | 2013-10-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
2NYX
 
 | | Crystal structure of RV1404 from Mycobacterium tuberculosis | | 分子名称: | Probable transcriptional regulatory protein, Rv1404 | | 著者 | Yu, M, Bursey, E.H, Radhakannan, R, Kim, C.-Y, Kaviratne, T, Woodruff, T, Segelke, B.W, Lekin, T, Toppani, D, Terwilliger, T.C, Hung, L.-W, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | 登録日 | 2006-11-21 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of RV1404 from Mycobacterium tuberculosis
To be Published
|
|
