3H8A
 
 | |
7BNL
 
 | | Notum ARUK3003710 | | 分子名称: | (4~{E})-2-(3,4-dimethylphenyl)-4-[(1-methylpyrazol-4-yl)methylidene]-1,3-oxazol-5-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y, Fish, P.V, Svensson, F, Steadman, D. | | 登録日 | 2021-01-22 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Notum Inhibitor ARUK3003710
To Be Published
|
|
3L6Y
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3L6X
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin, SULFATE ION | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
1IB6
 
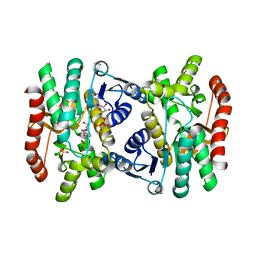 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | 分子名称: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | 登録日 | 2001-03-27 | | 公開日 | 2001-09-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1IE3
 
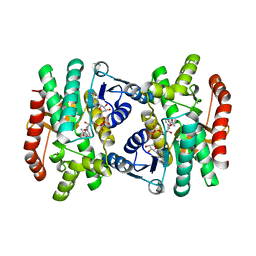 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | 分子名称: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | 著者 | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | 登録日 | 2001-04-05 | | 公開日 | 2001-09-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
7JNE
 
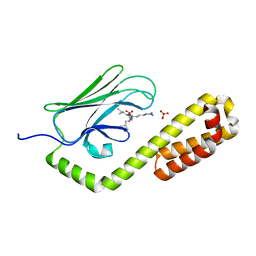 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RGSQLRIASR | | 分子名称: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | 著者 | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | 登録日 | 2020-08-04 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2JSX
 
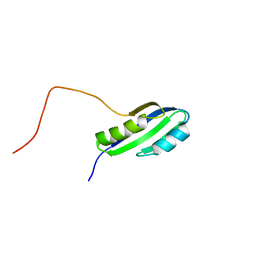 | |
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | 著者 | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | 登録日 | 2012-08-07 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
7JMM
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RAKNIILLSR | | 分子名称: | Alkaline phosphatase, Chaperone protein DnaK, SULFATE ION | | 著者 | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | 登録日 | 2020-08-02 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2I68
 
 | | Cryo-EM based theoretical model structure of transmembrane domain of the multidrug-resistance antiporter from E. coli EmrE | | 分子名称: | Protein emrE | | 著者 | Fleishman, S.J, Harrington, S.E, Enosh, A, Halperin, D, Tate, C.G, Ben-Tal, N. | | 登録日 | 2006-08-28 | | 公開日 | 2006-10-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (7.5 Å) | | 主引用文献 | Quasi-symmetry in the Cryo-EM Structure of EmrE Provides the Key to Modeling its Transmembrane Domain
J.Mol.Biol., 364, 2006
|
|
7B9H
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-42a | | 分子名称: | 1,2-ETHANEDIOL, 3-[(~{E})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ... | | 著者 | Torretta, A, Abbate, S, Parisini, E. | | 登録日 | 2020-12-14 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
4P6F
 
 | |
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | 分子名称: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-12 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
2O99
 
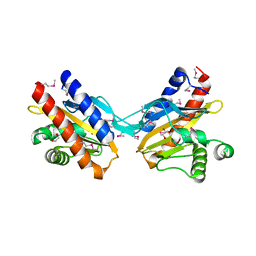 | | The crystal structure of E.coli IclR C-terminal fragment in complex with glyoxylate | | 分子名称: | 1,2-ETHANEDIOL, Acetate operon repressor, GLYCOLIC ACID | | 著者 | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | 登録日 | 2006-12-13 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
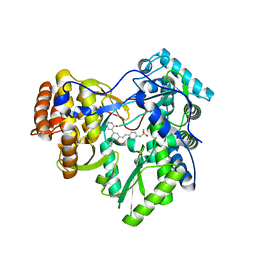
4MKB
 
 | |
4DL0
 
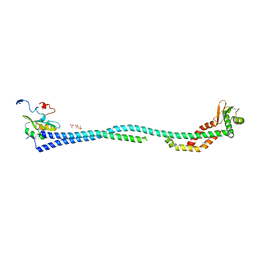 | | Crystal Structure of the heterotrimeric EGChead Peripheral Stalk Complex of the Yeast Vacuolar ATPase | | 分子名称: | SULFATE ION, TRIMETHYL LEAD ION, V-type proton ATPase subunit C, ... | | 著者 | Oot, R.A, Huang, L.S, Berry, E.A, Wilkens, S. | | 登録日 | 2012-02-05 | | 公開日 | 2012-10-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.905 Å) | | 主引用文献 | Crystal Structure of the Yeast Vacuolar ATPase Heterotrimeric EGC(head) Peripheral Stalk Complex.
Structure, 20, 2012
|
|
2O9A
 
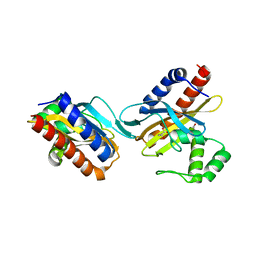 | | The crystal structure of the E.coli IclR C-terminal fragment in complex with pyruvate. | | 分子名称: | 1,2-ETHANEDIOL, Acetate operon repressor, PYRUVIC ACID | | 著者 | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | 登録日 | 2006-12-13 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
3S7H
 
 | | Structure of thrombin mutant Y225P in the E* form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin | | 著者 | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | 登録日 | 2011-05-26 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
1Y55
 
 | | Crystal structure of the C122S mutant of E. Coli expressed avidin related protein 4 (AVR4)-biotin complex | | 分子名称: | Avidin-related protein 4/5, BIOTIN, FORMIC ACID | | 著者 | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | 登録日 | 2004-12-02 | | 公開日 | 2005-05-24 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3ZJV
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | 分子名称: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | 著者 | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
4CDU
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | 分子名称: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-06 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
2WKX
 
 | | Crystal structure of the native E. coli zinc amidase AmiD | | 分子名称: | CHLORIDE ION, GLYCEROL, N-ACETYLMURAMOYL-L-ALANINE AMIDASE AMID, ... | | 著者 | Petrella, S, Kerff, F, Herman, R, Genereux, C, Pennartz, A, Sauvage, E, Joris, B, Charlier, P. | | 登録日 | 2009-06-18 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Specific Structural Features of the N-Acetylmuramoyl-L-Alanine Amidase Amid from Escherichia Coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
5B3A
 
 | | Crystal Structure of O-Phoshoserine Sulfhydrylase from Aeropyrum pernix in Complexed with the alpha-Aminoacrylate Intermediate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Protein CysO | | 著者 | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | 登録日 | 2016-02-12 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
7BAR
 
 | |
