4EWR
 
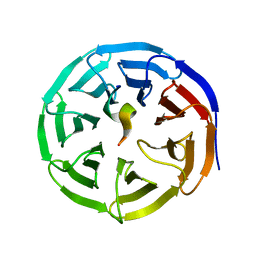 | | X-ray structure of WDR5-SETd1a Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-27 | | 公開日 | 2012-05-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
1V0P
 
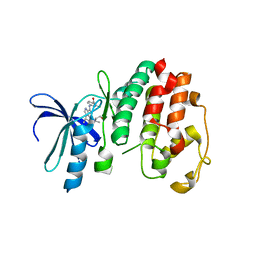 | | Structure of P. falciparum PfPK5-Purvalanol B ligand complex | | 分子名称: | CELL DIVISION CONTROL PROTEIN 2 HOMOLOG, PURVALANOL B | | 著者 | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | 登録日 | 2004-04-01 | | 公開日 | 2004-05-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
4ESG
 
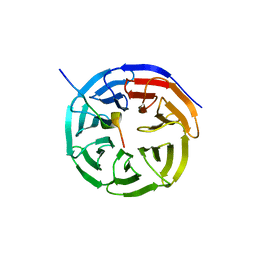 | | X-ray structure of WDR5-MLL1 Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase MLL, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-23 | | 公開日 | 2012-05-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-21 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
1TMF
 
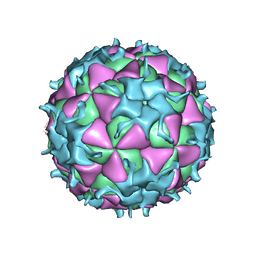 | |
4ERQ
 
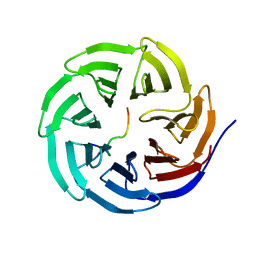 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-20 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.906 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
3PQK
 
 | |
3QAM
 
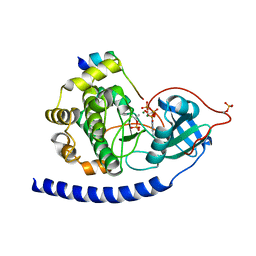 | | Crystal Structure of Glu208Ala mutant of catalytic subunit of cAMP-dependent protein kinase | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | 著者 | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | 登録日 | 2011-01-11 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
2JX8
 
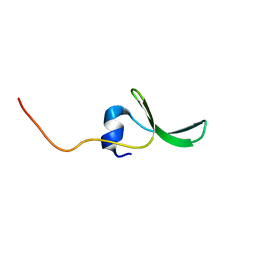 | | Solution structure of hPCIF1 WW domain | | 分子名称: | Phosphorylated CTD-interacting factor 1 | | 著者 | Kouno, T, Iwamoto, Y, Hirose, Y, Aizawa, T, Demura, M, Kawano, K, Ohkuma, Y, Mizuguchi, M. | | 登録日 | 2007-11-09 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1H, 13C, and 15N resonance assignments of hPCIF1 WW domain
To be Published
|
|
8HE7
 
 | | ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | 分子名称: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | 著者 | Wang, X.Y, Zhou, J, Xu, B.L. | | 登録日 | 2022-11-07 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
3L5X
 
 | | Crystal structure of the complex between IL-13 and H2L6 FAB | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, H2L6 HEAVY CHAIN, ... | | 著者 | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | 登録日 | 2009-12-22 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3L5W
 
 | | Crystal structure of the complex between IL-13 and C836 FAB | | 分子名称: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | 著者 | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | 登録日 | 2009-12-22 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
2RI7
 
 | |
6UCL
 
 | |
6UCI
 
 | | Transcription factor DeltaFosB bZIP domain self-assembly, oxidized form | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein fosB | | 著者 | Yin, Z, Machius, M, Rudenko, G. | | 登録日 | 2019-09-16 | | 公開日 | 2020-01-15 | | 最終更新日 | 2020-07-01 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
2YB4
 
 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound SO4, no metal | | 分子名称: | AMIDOHYDROLASE, SULFATE ION | | 著者 | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-03-01 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
1EXJ
 
 | | CRYSTAL STRUCTURE OF TRANSCRIPTION ACTIVATOR BMRR, FROM B. SUBTILIS, BOUND TO 21 BASE PAIR BMR OPERATOR AND TPP | | 分子名称: | DNA (5'-D(*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*T)-3'), MULTIDRUG-EFFLUX TRANSPORTER REGULATOR, SODIUM ION, ... | | 著者 | Zheleznova-Heldwein, E.E, Brennan, R.G. | | 登録日 | 2000-05-02 | | 公開日 | 2001-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of the transcription activator BmrR bound to DNA and a drug.
Nature, 409, 2001
|
|
8HLR
 
 | |
3QAL
 
 | | Crystal Structure of Arg280Ala mutant of Catalytic subunit of cAMP-dependent Protein Kinase | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | 著者 | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | 登録日 | 2011-01-11 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
1EXI
 
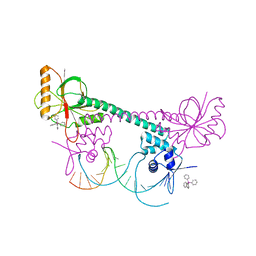 | | CRYSTAL STRUCTURE OF TRANSCRIPTION ACTIVATOR BMRR, FROM B. SUBTILIS, BOUND TO 21 BASE PAIR BMR OPERATOR AND TPSB | | 分子名称: | DNA (5'-D(*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*T)-3'), MULTIDRUG-EFFLUX TRANSPORTER REGULATOR, TETRAPHENYLANTIMONIUM ION, ... | | 著者 | Zheleznova-Heldwein, E.E, Brennan, R.G. | | 登録日 | 2000-05-02 | | 公開日 | 2001-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Crystal structure of the transcription activator BmrR bound to DNA and a drug.
Nature, 409, 2001
|
|
2YB1
 
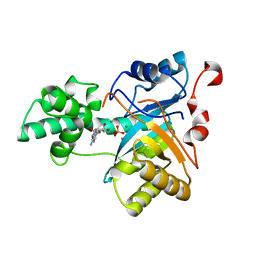 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound Mn, AMP and phosphate. | | 分子名称: | ADENOSINE MONOPHOSPHATE, AMIDOHYDROLASE, MANGANESE (II) ION, ... | | 著者 | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2011-02-25 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
4HPT
 
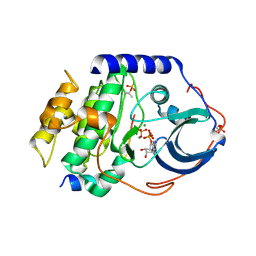 | | Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying complete phosphoryl transfer of AMP-PNP onto a substrate peptide | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | 著者 | Bastidas, A.C, Steichen, J.M, Wu, J, Taylor, S.S. | | 登録日 | 2012-10-24 | | 公開日 | 2013-03-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Phosphoryl transfer by protein kinase a is captured in a crystal lattice.
J.Am.Chem.Soc., 135, 2013
|
|
4HPU
 
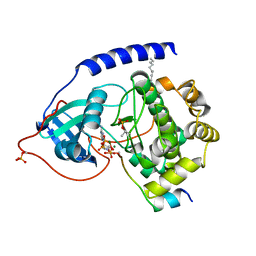 | | Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying partial phosphoryl transfer of AMP-PNP onto a substrate peptide | | 分子名称: | MAGNESIUM ION, MYRISTIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Bastidas, A.C, Steichen, J.M, Wu, J, Taylor, S.S. | | 登録日 | 2012-10-24 | | 公開日 | 2013-03-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Phosphoryl transfer by protein kinase a is captured in a crystal lattice.
J.Am.Chem.Soc., 135, 2013
|
|
5TCX
 
 | | Crystal structure of human tetraspanin CD81 | | 分子名称: | CD81 antigen, CHOLESTEROL | | 著者 | Zimmerman, B, McMillan, B.J, Seegar, T.C.M, Kruse, A.C, Blacklow, S.C. | | 登録日 | 2016-09-16 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.955 Å) | | 主引用文献 | Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket.
Cell, 167, 2016
|
|
2ZOQ
 
 | | Structural dissection of human mitogen-activated kinase ERK1 | | 分子名称: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 3, SODIUM ION, ... | | 著者 | Kinoshita, T, Tada, T, Nakae, S, Yoshida, I. | | 登録日 | 2008-06-01 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Crystal structure of human mono-phosphorylated ERK1 at Tyr204
Biochem.Biophys.Res.Commun., 377, 2008
|
|
