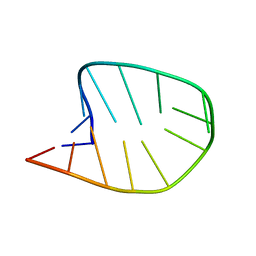
5SRP
 
 | |
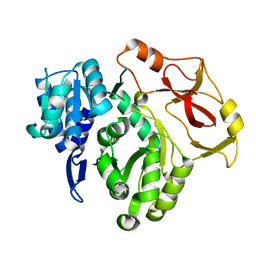
8HCZ
 
 | |
4V0N
 
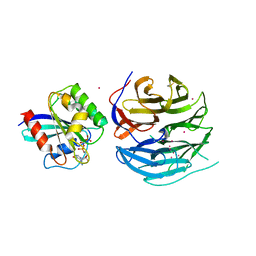 | |
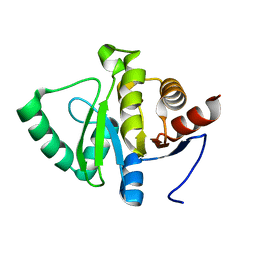
8CLR
 
 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | 分子名称: | RNA hairpin with CUUG tetraloop | | 著者 | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | 登録日 | 2023-02-17 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
5SRQ
 
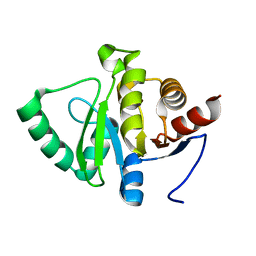 | |
8H70
 
 | |
4V1M
 
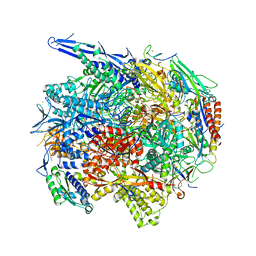 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | 分子名称: | 5'-D(*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP)-3', 5'-D(*CP*CP*AP*GP*GP*AP)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*CP *CP*BRUP*GP*GP*TP*C)-3', ... | | 著者 | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | 登録日 | 2014-09-29 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
5SRS
 
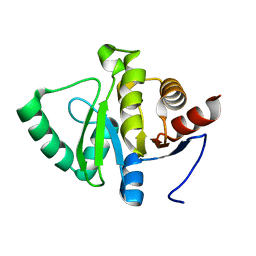 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z2614735107 - (R) and (S) isomers | | 分子名称: | 3-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one, 3-[(3S)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2RIW
 
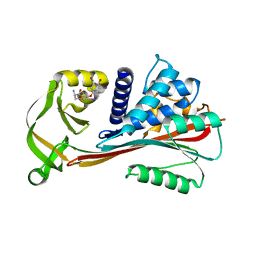 | | The Reactive loop cleaved human Thyroxine Binding Globulin complexed with thyroxine | | 分子名称: | 3,5,3',5'-TETRAIODO-L-THYRONINE, Thyroxine-binding globulin | | 著者 | Zhou, A, Wei, Z, Stanley, P.L.D, Read, R.J, Stein, P.E, Carrell, R.W. | | 登録日 | 2007-10-12 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Allosteric modulation of hormone release from thyroxine and corticosteroid-binding globulins
J.Biol.Chem., 286, 2011
|
|
8HDF
 
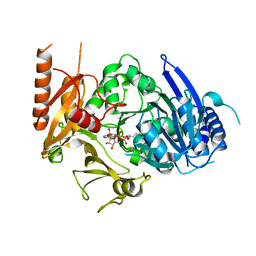 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | 分子名称: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | 登録日 | 2022-11-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
4UZN
 
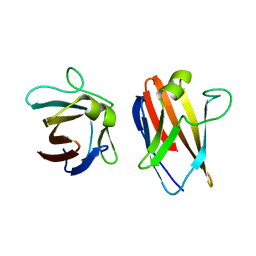 | | The native structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | 分子名称: | ENDO-BETA-1,4-GLUCANASE (CELULASE B) | | 著者 | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | 登録日 | 2014-09-05 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
5SP4
 
 | |
2RED
 
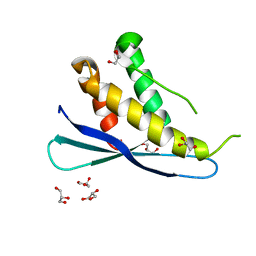 | | Crystal structures of C2ALPHA-PI3 kinase PX-domain domain indicate conformational change associated with ligand binding. | | 分子名称: | GLYCEROL, Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide | | 著者 | Parkinson, G.N, Vines, D, Driscoll, P.C, Djordjevic, S. | | 登録日 | 2007-09-26 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of PI3K-C2alpha PX domain indicate conformational change associated with ligand binding
Bmc Struct.Biol., 8, 2008
|
|
8HD4
 
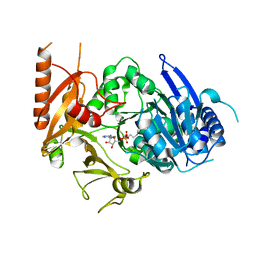 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | 分子名称: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | 著者 | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | 登録日 | 2022-11-03 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
2REK
 
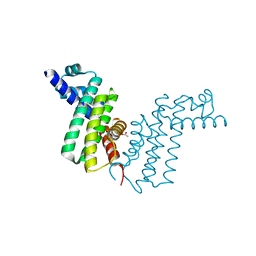 | | Crystal structure of tetR-family transcriptional regulator | | 分子名称: | ACETATE ION, Putative tetR-family transcriptional regulator | | 著者 | Dong, A, Xu, X, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-09-26 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of tetR-family transcriptional regulator.
To be Published
|
|
4V08
 
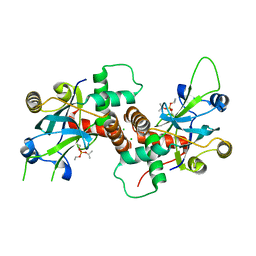 | | Inhibited dimeric pseudorabies virus protease pUL26N at 2 A resolution | | 分子名称: | CHLORIDE ION, DIISOPROPYL PHOSPHONATE, MAGNESIUM ION, ... | | 著者 | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | 登録日 | 2014-09-11 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
5SPC
 
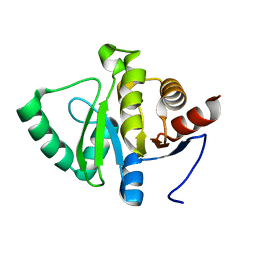 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894387 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HPJ
 
 | |
4W2O
 
 | |
5SRV
 
 | |
2RER
 
 | |
8GYI
 
 | | Crystal structure of Fic25 (holo form) from Streptomyces ficellus | | 分子名称: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE, ... | | 著者 | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | 登録日 | 2022-09-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
4V47
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | 分子名称: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | 登録日 | 2003-05-06 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (12.3 Å) | | 主引用文献 | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
5SRU
 
 | |
8HPK
 
 | | Crystal structure of the bacterial oxalate transporter OxlT in an oxalate-bound occluded form | | 分子名称: | Fab fragment Heavy chein, Fab fragment Light chain, OXALATE ION, ... | | 著者 | Shimamura, T, Hirai, T, Yamashita, A. | | 登録日 | 2022-12-12 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota.
Nat Commun, 14, 2023
|
|
