8IIL
 
 | |
8IIF
 
 | | H109A mutant of uracil DNA glycosylase X | | 分子名称: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIJ
 
 | | H109G mutant of uracil DNA glycosylase X | | 分子名称: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIP
 
 | |
3FC3
 
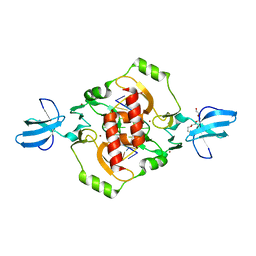 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I | | 分子名称: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', Restriction endonuclease Hpy99I, ... | | 著者 | Sokolowska, M, Czapinska, H, Bochtler, M. | | 登録日 | 2008-11-21 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
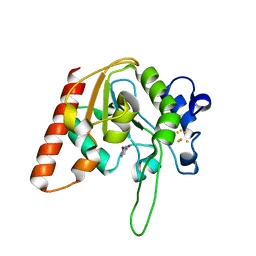
8IIN
 
 | |
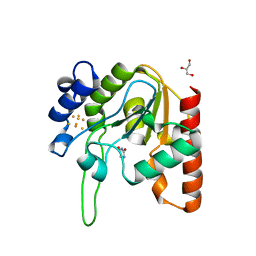
8III
 
 | |
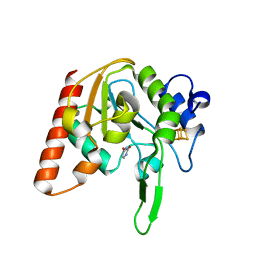
8IIG
 
 | |
8TG8
 
 | |
8D8M
 
 | |
1AA3
 
 | | C-TERMINAL DOMAIN OF THE E. COLI RECA, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | RECA | | 著者 | Aihara, H, Ito, Y, Kurumizaka, H, Terada, T, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1997-01-22 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-04-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing.
J.Mol.Biol., 274, 1997
|
|
6CC8
 
 | | Crystal structure MBD3 MBD domain in complex with methylated CpG DNA | | 分子名称: | Methyl-CpG-binding domain protein 3, UNKNOWN ATOM OR ION, methylated CpG DNA | | 著者 | Liu, K, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-06 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
7ODY
 
 | |
2FOK
 
 | |
6VR4
 
 | |
7ODX
 
 | |
4N6R
 
 | | Crystal structure of VosA-VelB-complex | | 分子名称: | SULFATE ION, VelB, VosA | | 著者 | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | 登録日 | 2013-10-14 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
4M35
 
 | | Crystal structure of gated-pore mutant H126/141D of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | 分子名称: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | 著者 | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | 登録日 | 2013-08-06 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
4M34
 
 | | Crystal structure of gated-pore mutant D138H of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | 分子名称: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | 著者 | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | 登録日 | 2013-08-06 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
5K8J
 
 | | Structure of Caulobacter crescentus VapBC1 (apo form) | | 分子名称: | GLYCEROL, Ribonuclease VapC, VapB family protein | | 著者 | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | 登録日 | 2016-05-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
5A77
 
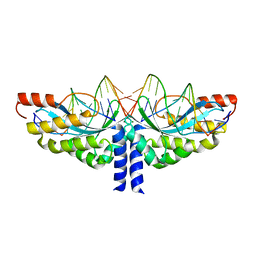 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA cleaved | | 分子名称: | 10MER DNA, 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP* GP*AP*DGP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', 14MER DNA, ... | | 著者 | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | 登録日 | 2015-07-03 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
3M9Q
 
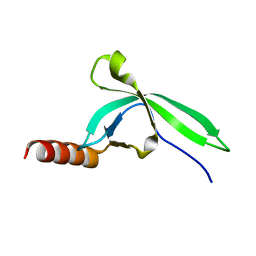 | |
6UIS
 
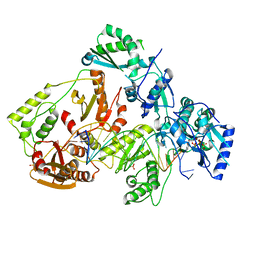 | | HIV-1 M184V reverse transcriptase-DNA complex with dCTP | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Primer DNA, ... | | 著者 | Lansdon, E.B. | | 登録日 | 2019-10-01 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.74833822 Å) | | 主引用文献 | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
6JG9
 
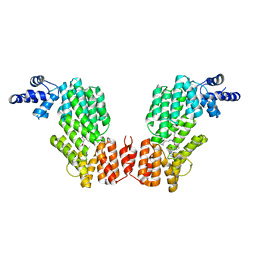 | |
6JG5
 
 | | Crystal structure of AimR | | 分子名称: | AimR transcriptional regulator | | 著者 | Guan, Z.Y, Pei, K, Zou, T.T. | | 登録日 | 2019-02-13 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.221 Å) | | 主引用文献 | Structural insights into DNA recognition by AimR of the arbitrium communication system in the SPbeta phage.
Cell Discov, 5, 2019
|
|
