7O6O
 
 | |
7NV4
 
 | |
7NXT
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-183 | | 分子名称: | 14-3-3 protein sigma, 4-(2,3-dihydro-1,4-benzoxazin-4-ylsulfonyl)benzaldehyde, CHLORIDE ION, ... | | 著者 | Wolter, M, Ottmann, C. | | 登録日 | 2021-03-19 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NZK
 
 | |
7O3Q
 
 | |
7AXZ
 
 | | Ku70/80 complex apo form | | 分子名称: | X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | 著者 | Hnizda, A, Tesina, P, Novak, P, Blundell, T.L. | | 登録日 | 2020-11-10 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | SAP domain forms a flexible part of DNA aperture in Ku70/80.
Febs J., 288, 2021
|
|
7O5A
 
 | |
7O6G
 
 | |
7AH4
 
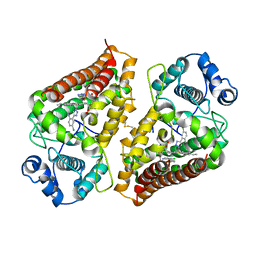 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0363 | | 分子名称: | 4-chloranyl-2-(2~{H}-1,2,3-triazol-4-yl)aniline, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | 登録日 | 2020-09-24 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AH6
 
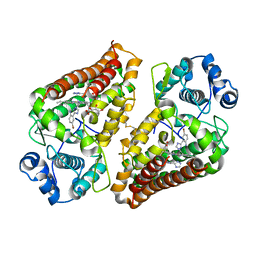 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0752 | | 分子名称: | 4-bromanyl-2-(4~{H}-1,2,4-triazol-3-yl)aniline, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | 著者 | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | 登録日 | 2020-09-24 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.998 Å) | | 主引用文献 | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AQF
 
 | |
5AZG
 
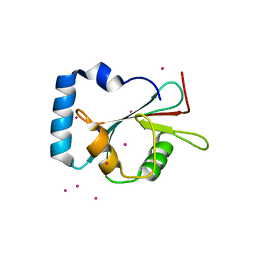 | | Crystal structure of LGG-1 complexed with a UNC-51 peptide | | 分子名称: | CADMIUM ION, Protein lgg-1, Serine/threonine-protein kinase unc-51 | | 著者 | Watanabe, Y, Fujioka, Y, Noda, N.N. | | 登録日 | 2015-10-05 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
1QP8
 
 | |
7AH5
 
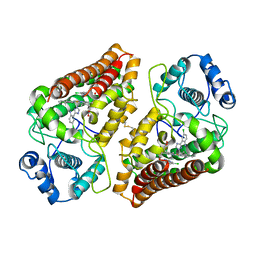 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0706 | | 分子名称: | 4-chloranyl-2-(1~{H}-1,2,4-triazol-5-yl)aniline, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | 登録日 | 2020-09-24 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AQG
 
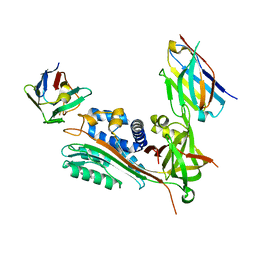 | | Crystal Structure of Small Molecule Inhibitor TM5484 Bound to Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) | | 分子名称: | 5-Chloro-2-[[2-[3-(furan-3-yl)anilino]-2-oxoacetyl]amino]benzoic acid, Plasminogen activator inhibitor 1, VHH-2g-42 (Nb42), ... | | 著者 | Sillen, M, Strelkov, S.V, Declerck, P.J. | | 登録日 | 2020-10-21 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Structural Insight into the Two-Step Mechanism of PAI-1 Inhibition by Small Molecule TM5484.
Int J Mol Sci, 22, 2021
|
|
1Q6D
 
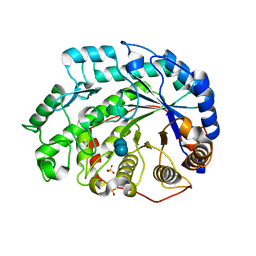 | | Crystal structure of Soybean Beta-Amylase Mutant (M51T) with Increased pH Optimum | | 分子名称: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | 著者 | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | 登録日 | 2003-08-13 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
7B24
 
 | |
7O24
 
 | |
7B1Y
 
 | |
5AWO
 
 | | Arthrobacter globiformis T6 isomalto-dextranse | | 分子名称: | ACETATE ION, Isomaltodextranase, PHOSPHATE ION | | 著者 | Tonozuka, T. | | 登録日 | 2015-07-08 | | 公開日 | 2015-09-09 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Crystal Structure and Mutational Analysis of Isomalto-dextranase, a Member of Glycoside Hydrolase Family 27
J.Biol.Chem., 290, 2015
|
|
7B23
 
 | |
7O0H
 
 | |
7B20
 
 | |
5AZH
 
 | | Crystal structure of LGG-2 fused with an EEEWEEL peptide | | 分子名称: | EEEWEEL peptide,Protein lgg-2, MAGNESIUM ION | | 著者 | Watanabe, Y, Fujioka, Y, Noda, N.N. | | 登録日 | 2015-10-05 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
7O31
 
 | | Crystal structure of the anti-PAS Fab 1.2 in complex with its epitope peptide and the anti-Kappa VHH domain | | 分子名称: | 1,2-ETHANEDIOL, PAS#1 epitope peptide, anti-Kappa VHH domain, ... | | 著者 | Schilz, J, Schiefner, A, Skerra, A. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
