8HVX
 
 | |
6W5J
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7d | | 分子名称: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
5ZGE
 
 | | Crystal structure of NDM-1 at pH5.5 (Bis-Tris) in complex with hydrolyzed ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | 著者 | Zhang, H, Hao, Q. | | 登録日 | 2018-03-08 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGR
 
 | | Crystal structure of NDM-1 at pH7.3 (HEPES) in complex with hydrolyzed ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | 著者 | Zhang, H, Hao, Q. | | 登録日 | 2018-03-10 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
6VKD
 
 | | Crystal Structure of Inhibitor JNJ-36689282 in Complex with Prefusion RSV F Glycoprotein | | 分子名称: | 1-cyclopropyl-3-({1-[3-(methylsulfonyl)propyl]-1H-pyrrolo[3,2-c]pyridin-2-yl}methyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one, CHLORIDE ION, Prefusion RSV F (DS-Cav1), ... | | 著者 | McLellan, J.S. | | 登録日 | 2020-01-20 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
5ZGV
 
 | |
8HVY
 
 | |
8HVZ
 
 | |
8HVW
 
 | |
7AYN
 
 | | Crystal structure of the lectin domain of the FimH variant Arg98Ala, in complex with Methyl 3-chloro-4-D-mannopyranosyloxy-3-biphenylcarboxylate | | 分子名称: | 1,2-ETHANEDIOL, Type 1 fimbrin D-mannose specific adhesin, methyl 3-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]benzoate | | 著者 | Jakob, R.P, Tomasic, T, Rabbani, S, Reisner, A, Jakopin, Z, Maier, T, Ernst, B, Anderluh, M. | | 登録日 | 2020-11-12 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Does targeting Arg98 of FimH lead to high affinity antagonists?
Eur.J.Med.Chem., 211, 2020
|
|
7B3O
 
 | |
8FRW
 
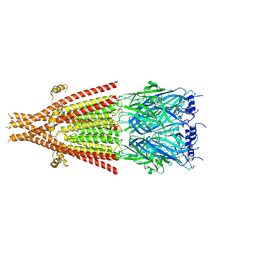 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, pre-activated | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | 著者 | Felt, K.C, Chakrapani, S. | | 登録日 | 2023-01-09 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FRX
 
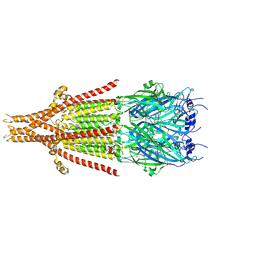 | | Full-length mouse 5-HT3A receptor in complex with SMP100, pre-activated | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | 著者 | Felt, K.C, Chakrapani, S. | | 登録日 | 2023-01-09 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4H1Y
 
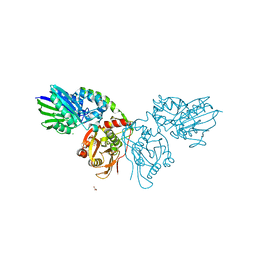 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with PSB11552 | | 分子名称: | 2,2'-[(2-{[2-({[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)ethyl]amino}-2-oxoethyl)imino]diacetic acid (non-preferred name), 5'-nucleotidase, CALCIUM ION, ... | | 著者 | Pippel, J, Zebisch, M, Knapp, K, Straeter, N. | | 登録日 | 2012-09-11 | | 公開日 | 2012-11-28 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
5U55
 
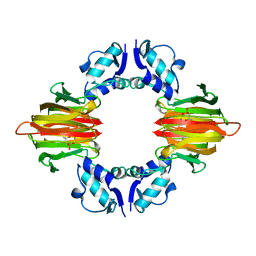 | | Psf4 in complex with Mn2+ and (S)-2-HPP | | 分子名称: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, (S)-2-hydroxypropylphosphonic acid epoxidase, MANGANESE (II) ION | | 著者 | Chekan, J.R, Nair, S.K. | | 登録日 | 2016-12-06 | | 公開日 | 2017-01-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
7AG8
 
 | |
5U5D
 
 | | Psf4 in complex with Mn2+ and (R)-2-HPP | | 分子名称: | (S)-2-hydroxypropylphosphonic acid epoxidase, MANGANESE (II) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | 著者 | Chekan, J.R, Nair, S.K. | | 登録日 | 2016-12-06 | | 公開日 | 2017-01-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
8FSZ
 
 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, open-like | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | 著者 | Felt, K.C, Chakrapani, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.79 Å) | | 主引用文献 | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7USZ
 
 | | Human DDAH-1, holo (Zn-bound) form | | 分子名称: | CHLORIDE ION, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, ZINC ION | | 著者 | Smith, C.A, Ghebre, Y.T. | | 登録日 | 2022-04-26 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
5YUP
 
 | | Crystal Structure of the Fab fragment of FVIIa antibody mAb4F5 | | 分子名称: | the heavy chain of the Fab fragment of FVIIa antibody mAb4F5, the light chain of the Fab fragment of FVIIa antibody mAb4F5 | | 著者 | Jiang, L.G, Persson, E, Huang, M.D. | | 登録日 | 2017-11-22 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystal structure, epitope, and functional impact of an antibody against a superactive FVIIa provide insights into allosteric mechanism.
Res Pract Thromb Haemost, 3, 2019
|
|
5TOZ
 
 | | JAK3 with covalent inhibitor PF-06651600 | | 分子名称: | 1-{(2S,5R)-2-methyl-5-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}propan-1-one, SULFATE ION, Tyrosine-protein kinase JAK3 | | 著者 | Vajdos, F.F. | | 登録日 | 2016-10-19 | | 公開日 | 2016-11-09 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Discovery of a JAK3-Selective Inhibitor: Functional Differentiation of JAK3-Selective Inhibition over pan-JAK or JAK1-Selective Inhibition.
ACS Chem. Biol., 11, 2016
|
|
7B3C
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -4 (structure 2) | | 分子名称: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | 登録日 | 2020-11-30 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
5YVO
 
 | | Human Glutathione Transferase Omega1 covalently bound to ML175 inhibitor | | 分子名称: | ACETATE ION, Glutathione S-transferase omega-1, N-{3-[(2-chloro-acetyl)-(4-nitro-phenyl)-amino]-propyl}-2,2,2-trifluoro-acetamide, ... | | 著者 | Saisawang, C, Ketterman, A, Wongsantichon, J. | | 登録日 | 2017-11-27 | | 公開日 | 2018-11-28 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
7UT0
 
 | | Human DDAH-1, apo form | | 分子名称: | 1,2-ETHANEDIOL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | 著者 | Smith, C.A, Ghebre, Y.T. | | 登録日 | 2022-04-26 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7UW0
 
 | | Crystal structure of human ClpP protease in complex with TR-133 | | 分子名称: | 3-({3-[(4-bromophenyl)methyl]-4-oxo-3,5,7,8-tetrahydropyrido[4,3-d]pyrimidin-6(4H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | 著者 | Mabanglo, M.F, Houry, W.A. | | 登録日 | 2022-05-02 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
