8FOI
 
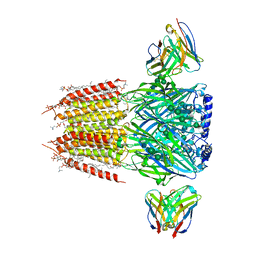 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, DODECANE, ... | | 著者 | Sun, C, Gouaux, E. | | 登録日 | 2022-12-30 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
9EMV
 
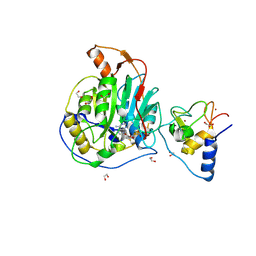 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | 登録日 | 2024-03-11 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
6OPU
 
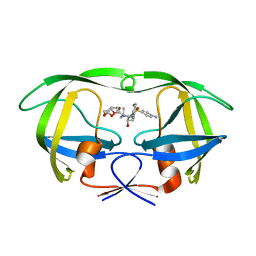 | | HIV-1 Protease NL4-3 K45I, M46I, V82F, I84V Mutant in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2019-04-25 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPY
 
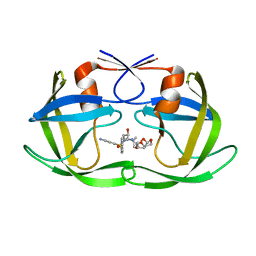 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, A71V, L76V, V82F, I84V Mutant in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2019-04-25 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
8XOI
 
 | | Cryo-EM structure of GPR30-Gq complex structure in the presence of fulvestrant | | 分子名称: | G-protein coupled estrogen receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, H, Xu, P, Xu, H.E. | | 登録日 | 2024-01-01 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and functional evidence that GPR30 is not a direct estrogen receptor.
Cell Res., 34, 2024
|
|
8X8N
 
 | | Cryo-EM structure of the octreotide-bound Somatostatin receptor 5-Gi protein complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha, ... | | 著者 | Xu, H.E, You, C, Zhao, L, Li, J. | | 登録日 | 2023-11-27 | | 公開日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structure of the octreotide-bound Somatostatin receptor 5-Gi protein complex
To Be Published
|
|
8FLJ
 
 | | Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA | | 分子名称: | CRISPR leader and repeat, anti-sense strand of DNA, CRISPR leader, ... | | 著者 | Santiago-Frangos, A, Henriques, W.S, Wiegand, T, Gauvin, C, Buyukyoruk, M, Neselu, K, Eng, E.T, Lander, G.C, Wiedenheft, B. | | 登録日 | 2022-12-21 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8QCF
 
 | | yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Antiviral helicase SKI2, Exosome complex component CSL4, ... | | 著者 | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | 登録日 | 2023-08-25 | | 公開日 | 2024-01-10 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8X8L
 
 | | Cryo-EM structure of the cortistatin 17-bound Somatostatin receptor 5-Gi protein complex | | 分子名称: | Cortistatin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, H.E, You, C, Zhao, L, Li, J. | | 登録日 | 2023-11-27 | | 公開日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structure of the cortistatin 17-bound Somatostatin receptor 5-Gi protein complex
To Be Published
|
|
7NMB
 
 | |
8FIM
 
 | | Structure of APOBEC3A (E72A inactive mutant) in complex with TTC-hairpin DNA substrate | | 分子名称: | CHLORIDE ION, DNA (5'-D(*TP*GP*CP*GP*CP*TP*TP*CP*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, ... | | 著者 | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | 登録日 | 2022-12-16 | | 公開日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | DNA-based inhibitors restrict mutagenic activity of APOBEC3 in cells
To Be Published
|
|
6S0T
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, sulfate, soaked with iodide | | 分子名称: | IODIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | 著者 | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
8A41
 
 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | 分子名称: | p70 | | 著者 | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | 登録日 | 2022-06-10 | | 公開日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (4.88 Å) | | 主引用文献 | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
6UQK
 
 | | Cryo-EM structure of type 3 IP3 receptor revealing presence of a self-binding peptide | | 分子名称: | ZINC ION, inositol 1,4,5-triphosphate receptor, type 3 | | 著者 | Azumaya, C.M, Linton, E.A, Risener, C.J, Nakagawa, T, Karakas, E. | | 登録日 | 2019-10-20 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Cryo-EM structure of human type-3 inositol triphosphate receptor reveals the presence of a self-binding peptide that acts as an antagonist.
J.Biol.Chem., 295, 2020
|
|
7E1J
 
 | |
6XKE
 
 | |
6XL7
 
 | |
6I8B
 
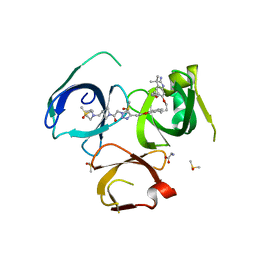 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | 分子名称: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | 著者 | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | 登録日 | 2018-11-19 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
8A3C
 
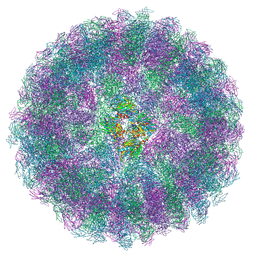 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | 分子名称: | p70 | | 著者 | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | 登録日 | 2022-06-08 | | 公開日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A6J
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | | 分子名称: | p70 | | 著者 | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | 登録日 | 2022-06-17 | | 公開日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
1E74
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R11E | | 分子名称: | ALPHA-CONOTOXIN IM1(R11E) | | 著者 | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | 登録日 | 2000-08-24 | | 公開日 | 2000-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E76
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT D5N | | 分子名称: | ALPHA-CONOTOXIN IM1(D5N) | | 著者 | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | 登録日 | 2000-08-24 | | 公開日 | 2000-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E75
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R7L | | 分子名称: | ALPHA-CONOTOXIN IM1(R7L) | | 著者 | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | 登録日 | 2000-08-24 | | 公開日 | 2000-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1WUT
 
 | | Acyl Ureas as Human Liver Glycogen Phosphorylase Inhibitors for the Treatment of Type 2 Diabetes | | 分子名称: | 7-[2,6-DICHLORO-4-({[(2-CHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]HEPTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | 著者 | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.-J, Herling, A.W, Oikonomakos, N.G, Kosmopoulou, M.N, Schmoll, D, Sarubbi, E, von Roedern, E, Schonafinger, K, Defossa, E. | | 登録日 | 2004-12-08 | | 公開日 | 2005-12-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
5IJ0
 
 | | Cryo EM density of microtubule assembled from human TUBB3 | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Ti, S.-C, Pamula, M.C, Howes, S.C, Duellberg, C, Nicholas, C.I, Kleiner, R.E, Forth, S, Surrey, T, Nogales, E, Kapoor, T.M. | | 登録日 | 2016-03-01 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Mutations in Human Tubulin Proximal to the Kinesin-Binding Site Alter Dynamic Instability at Microtubule Plus- and Minus-Ends.
Dev.Cell, 37, 2016
|
|
