5KB3
 
 | |
8P7Q
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7P5I
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 1-[3-[3-(dimethylcarbamoyl)phenyl]phenyl]-5-[(1~{R},2~{R})-2-(1-methyl-1,2,3-triazol-4-yl)cyclopropyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | 著者 | Davies, T.G, Cleasby, A. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6YM4
 
 | | Crystal structure of BAY-297 with PIP4K2A | | 分子名称: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | 著者 | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | 登録日 | 2020-04-07 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
8ZD2
 
 | | NMR structure of the (CGG-dsDNA:ND=) 1:2 complex | | 分子名称: | DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3'), ~{N}-(7-methyl-1,8-naphthyridin-2-yl)-3-[[3-[(7-methyl-1,8-naphthyridin-2-yl)amino]-3-oxidanylidene-propyl]amino]propanamide | | 著者 | Sakurabayashi, S, Furuita, K, Yamada, T, Nomura, M, Nakatani, K, Kojima, C. | | 登録日 | 2024-05-01 | | 公開日 | 2025-04-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the (CGG-dsDNA:ND=) 1:2 complex
To Be Published
|
|
6YSJ
 
 | | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-(4-bromophenyl)ethanone, DIMETHYL SULFOXIDE, ... | | 著者 | Scanlan, W, Heine, A, Klebe, G, Abazi, N, Paulus, A. | | 登録日 | 2020-04-22 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10)
To be published
|
|
7NEG
 
 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody COVOX-269 Fab heavy chain, Antibody COVOX-269 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-02-04 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera.
Cell, 184, 2021
|
|
6YBE
 
 | |
6X4Q
 
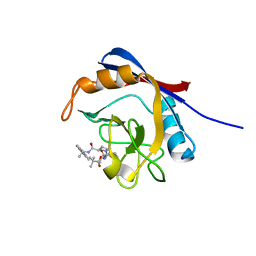 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 33) | | 分子名称: | (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | 登録日 | 2020-05-22 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6Y3K
 
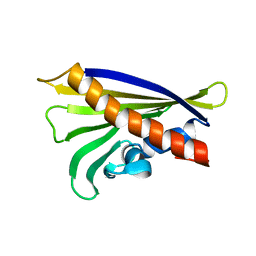 | | NMR solution structure of the hazelnut allergen Cor a 1.0403 | | 分子名称: | Major allergen variant Cor a 1.0403 | | 著者 | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | 登録日 | 2020-02-18 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
7NTH
 
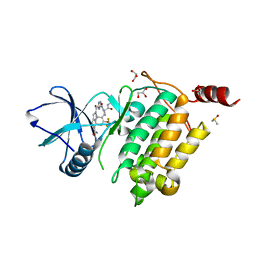 | | Structure of TAK1 in complex with compound 54 | | 分子名称: | 2-[[5-[[2-[bis(fluoranyl)methoxy]phenyl]methyl-[(2~{R})-1-(methylamino)-1-oxidanylidene-propan-2-yl]carbamoyl]-1~{H}-imidazol-2-yl]carbonyl]isoindole-5-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | 登録日 | 2021-03-09 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
8ZD3
 
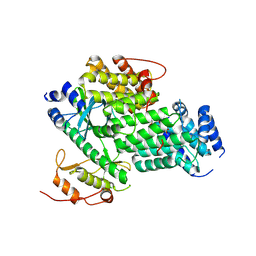 | | Crystal structure of ALPK1-N+K in complex with CDP-heptose | | 分子名称: | (2R,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]oxane-2,3,4,5-tetrol, Alpha-protein kinase 1, CYTIDINE-5'-DIPHOSPHATE, ... | | 著者 | She, Y, Ding, J.J, Shao, F. | | 登録日 | 2024-05-01 | | 公開日 | 2025-05-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | crystal structure of Alpha-kinase 1 N+K in complex with CDP-hep
To be published
|
|
7NXA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 B.1.351 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
5MTX
 
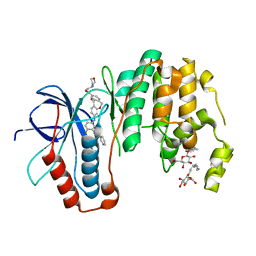 | | Dibenzooxepinone inhibitor 12b in complex with p38 MAPK | | 分子名称: | 3-[(3-benzamido-4-fluoranyl-phenyl)amino]-~{N}-(2-morpholin-4-ylethyl)-11-oxidanylidene-6~{H}-benzo[c][1]benzoxepine-9-carboxamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | 著者 | Buehrmann, M, Rauh, D. | | 登録日 | 2017-01-11 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of Novel Type I(1)/2 p38 alpha MAP Kinase Inhibitors with Excellent Selectivity, High Potency, and Prolonged Target Residence Time by Interfering with the R-Spine.
J. Med. Chem., 60, 2017
|
|
7NXB
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 P.1 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
8OMK
 
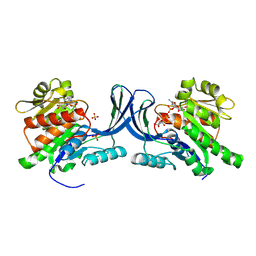 | | hKHK-C in complex with ADP & fructose 1-phosphate | | 分子名称: | 1-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, ... | | 著者 | Ebenhoch, R, Pautsch, A. | | 登録日 | 2023-03-31 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
8ZEP
 
 | |
8J3O
 
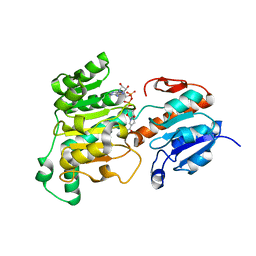 | | Formate dehydrogenase wild-type enzyme from Candida dubliniensis complexed with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Formate dehydrogenase, MAGNESIUM ION | | 著者 | Ma, W, Zheng, Y.C, Geng, Q, Chen, C. | | 登録日 | 2023-04-17 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Engineering a Formate Dehydrogenase for NADPH Regeneration.
Chembiochem, 24, 2023
|
|
8TUK
 
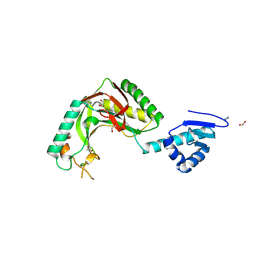 | | Alvinella ASCC1 KH and Phosphodiesterase/Ligase Domain | | 分子名称: | 1,2-ETHANEDIOL, Activating signal cointegrator 1 complex subunit 1, IMIDAZOLE | | 著者 | Tsutakawa, S.E, Tainer, J.A, Arvai, A.S, Chinnam, N.B. | | 登録日 | 2023-08-16 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | ASCC1 structures and bioinformatics reveal a novel helix-clasp-helix RNA-binding motif linked to a two-histidine phosphodiesterase.
J.Biol.Chem., 300, 2024
|
|
8ZEO
 
 | |
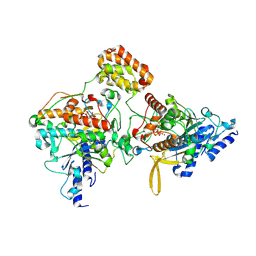
8ZEN
 
 | |
5MTY
 
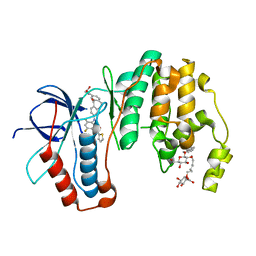 | | Dibenzosuberone inhibitor 8e in complex with p38 MAPK | | 分子名称: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside, ~{N}-[2,4-bis(fluoranyl)-5-[[14-(2-hydroxyethylcarbamoyl)-2-oxidanylidene-6-tricyclo[9.4.0.0^{3,8}]pentadeca-1(15),3(8),4,6,11,13-hexaenyl]amino]phenyl]thiophene-2-carboxamide | | 著者 | Buehrmann, M, Rauh, D. | | 登録日 | 2017-01-11 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of Novel Type I(1)/2 p38 alpha MAP Kinase Inhibitors with Excellent Selectivity, High Potency, and Prolonged Target Residence Time by Interfering with the R-Spine.
J. Med. Chem., 60, 2017
|
|
8AV0
 
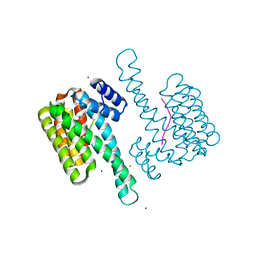 | | small molecule stabilizer (compound 1) for C-RAF pS259 and 14-3-3 | | 分子名称: | 1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-4-carboxamide, 14-3-3 protein sigma, CHLORIDE ION, ... | | 著者 | Visser, E.J, Sijbesma, E, Ottmann, C. | | 登録日 | 2022-08-26 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
6OGT
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-001 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
5MMN
 
 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea | | 分子名称: | 1-ethyl-3-[8-methyl-5-(2-methylpyridin-4-yl)isoquinolin-3-yl]urea, DNA gyrase subunit B | | 著者 | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
