6KAO
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 95 K: Dark | | 分子名称: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | 著者 | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | 登録日 | 2019-06-23 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAR
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 140 K: Light | | 分子名称: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | 著者 | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | 登録日 | 2019-06-24 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAH
 
 | | Crosslinked alpha(Ni)-beta(Fe-CO) human hemoglobin A in the T quaternary structure at 95 K: Dark | | 分子名称: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | 著者 | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | 登録日 | 2019-06-23 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAQ
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 140 K: Dark | | 分子名称: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | 著者 | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | 登録日 | 2019-06-24 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAU
 
 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 140 K: Dark | | 分子名称: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | 著者 | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | 登録日 | 2019-06-24 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1R43
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri (selenomethionine substituted protein) | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-AMINO ISOBUTYRATE, ... | | 著者 | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | 登録日 | 2003-10-03 | | 公開日 | 2003-11-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278
|
|
4OZ1
 
 | |
6KA9
 
 | | Crosslinked alpha(Fe-CO)-beta(Ni) human hemoglobin A in the T quaternary structure at 95 K: Dark | | 分子名称: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | 著者 | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | 登録日 | 2019-06-21 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6M1I
 
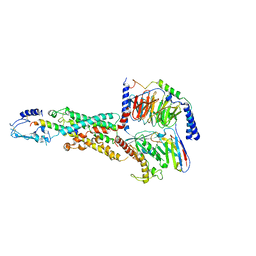 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | 登録日 | 2020-02-26 | | 公開日 | 2020-03-11 | | 最終更新日 | 2020-05-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
1BNB
 
 | |
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | 分子名称: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | 著者 | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | 登録日 | 2017-11-07 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
1NER
 
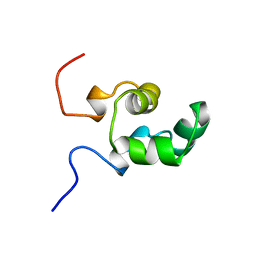 | |
4ZTH
 
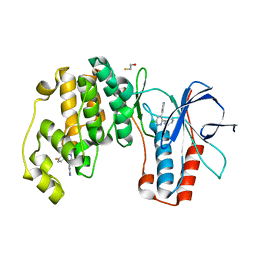 | | Structure of human p38aMAPK-arylpyridazinylpyridine fragment complex used in inhibitor discovery | | 分子名称: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, 6-chloro-3-phenyl-4-(pyridin-4-yl)pyridazine, BETA-MERCAPTOETHANOL, ... | | 著者 | Grum-Tokars, V.L, Roy, S.M, Watterson, D.M. | | 登録日 | 2015-05-14 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A Selective and Brain Penetrant p38 alpha MAPK Inhibitor Candidate for Neurologic and Neuropsychiatric Disorders That Attenuates Neuroinflammation and Cognitive Dysfunction.
J.Med.Chem., 62, 2019
|
|
1DAQ
 
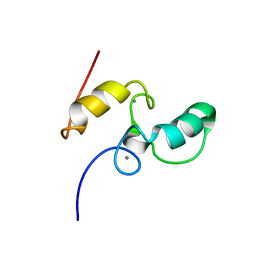 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (MINIMIZED AVERAGE STRUCTURE) | | 分子名称: | CALCIUM ION, ENDOGLUCANASE SS | | 著者 | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | 登録日 | 1999-10-31 | | 公開日 | 2001-04-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
4QJZ
 
 | | Pneumocystis carinii dihydrofolate reductase ternary complex with NADPH and the inhibitor 24, (N~6~-METHYL-N~6~-(NAPHTHALEN-1-YL)PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | 分子名称: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-(naphthalen-1-yl)pyrido[2,3-d]pyrimidine-2,4,6-triamine | | 著者 | Cody, V. | | 登録日 | 2014-06-05 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4Q5V
 
 | | Crystal structure of the catalytic core of human DNA polymerase alpha in ternary complex with an RNA-primed DNA template and aphidicolin | | 分子名称: | (3R,4R,4aR,6aS,8R,9R,11aS,11bS)-4,9-bis(hydroxymethyl)-4,11b-dimethyltetradecahydro-8,11a-methanocyclohepta[a]naphthale ne-3,9-diol, DNA polymerase alpha catalytic subunit, DNA template, ... | | 著者 | Baranovskiy, A.G, Babayeva, N.D, Suwa, Y, Gu, J, Tahirov, T.H. | | 登録日 | 2014-04-17 | | 公開日 | 2014-11-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Structural basis for inhibition of DNA replication by aphidicolin.
Nucleic Acids Res., 42, 2014
|
|
1HSL
 
 | |
1ELT
 
 | |
6MUT
 
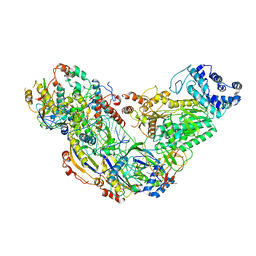 | | Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | | 分子名称: | RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*GP*C)-3'), RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP*C)-3'), Uncharacterized protein Csm1, ... | | 著者 | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | 登録日 | 2018-10-23 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
4QJC
 
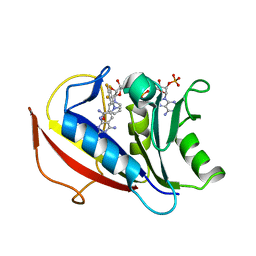 | | Human dihydrofolate reductase ternary complex with NADPH and inhibitor 26 (N~6~-METHYL-N~6~-(3,4,5-TRIFLUOROPHENYL)PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | 分子名称: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-(3,4,5-trifluorophenyl)pyrido[2,3-d]pyrimidine-2,4,6-triamine | | 著者 | Cody, V. | | 登録日 | 2014-06-03 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
1VAS
 
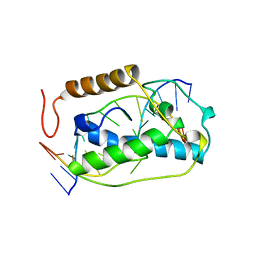 | | ATOMIC MODEL OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME COMPLEXED WITH A DNA SUBSTRATE: STRUCTURAL BASIS FOR DAMAGED DNA RECOGNITION | | 分子名称: | DNA (5'-D(*AP*TP*CP*GP*CP*GP*TP*TP*GP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*GP*A)-3'), PROTEIN (T4 ENDONUCLEASE V (E.C.3.1.25.1)) | | 著者 | Vassylyev, D.G, Kashiwagi, T, Mikami, Y, Ariyoshi, M, Iwai, S, Ohtsuka, E, Morikawa, K. | | 登録日 | 1995-09-08 | | 公開日 | 1996-01-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.
Cell(Cambridge,Mass.), 83, 1995
|
|
1MUT
 
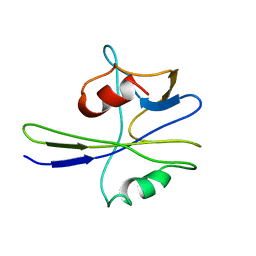 | | NMR STUDY OF MUTT ENZYME, A NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | 分子名称: | NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | 著者 | Abeygunawardana, C, Weber, D.J, Gittis, A.G, Frick, D.N, Lin, J, Miller, A.-F, Bessman, M.J, Mildvan, A.S. | | 登録日 | 1995-09-14 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase.
Biochemistry, 34, 1995
|
|
2JPH
 
 | |
2PTL
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN-BINDING DOMAIN OF PROTEIN L. COMPARISON WITH THE IGG-BINDING DOMAINS OF PROTEIN G | | 分子名称: | PROTEIN L | | 著者 | Wikstroem, M, Drakenberg, T, Forsen, S, Sjoebring, U, Bjoerck, L. | | 登録日 | 1994-08-12 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G.
Biochemistry, 33, 1994
|
|
3OBL
 
 | |
