1NLA
 
 | | Solution Structure of Switch Arc, a Mutant with 3(10) Helices Replacing a Wild-Type Beta-Ribbon | | 分子名称: | Transcriptional repressor arc | | 著者 | Cordes, M.H, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | 登録日 | 2003-01-06 | | 公開日 | 2003-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Switch Arc, a mutant with 3(10) helices replacing a wild-type beta-ribbon
J.Mol.Biol., 326, 2003
|
|
6SVE
 
 | | Protein allostery of the WW domain at atomic resolution: pCdc25C bound structure | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | 登録日 | 2019-09-18 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7AEP
 
 | | Solution structure of U1-A RRM2 (190-282) | | 分子名称: | U1 small nuclear ribonucleoprotein A | | 著者 | Campagne, S, Allain, F.H. | | 登録日 | 2020-09-18 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An in vitro reconstituted U1 snRNP allows the study of the disordered regions of the particle and the interactions with proteins and ligands.
Nucleic Acids Res., 49, 2021
|
|
6XB7
 
 | | IRES-targeting Small Molecule Inhibits Enterovirus 71 Replication via Allosteric Stabilization of a Ternary Complex | | 分子名称: | 3-amino-N-(diaminomethylidene)-5-(dimethylamino)-6-(phenylethynyl)pyrazine-2-carboxamide, RNA (41-MER) | | 著者 | Tolbert, B.S, Davila, J, Sugarman, A.L, Chiu, L.Y. | | 登録日 | 2020-06-05 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex.
Nat Commun, 11, 2020
|
|
6SVC
 
 | | Protein allostery of the WW domain at atomic resolution: apo structure | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | 登録日 | 2019-09-18 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6N8F
 
 | |
8PEK
 
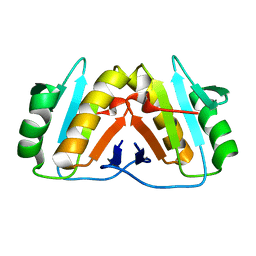 | |
3FT7
 
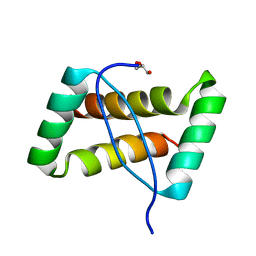 | | Crystal structure of an extremely stable dimeric protein from sulfolobus islandicus | | 分子名称: | GLYCEROL, Uncharacterized protein ORF56 | | 著者 | Neumann, P, Loew, C, Weininger, U, Stubbs, M.T. | | 登録日 | 2009-01-12 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Stability Analysis of an Extremely Stable Dimeric DNA Binding Protein from Sulfolobus islandicus
Biochemistry, 48, 2009
|
|
3F0P
 
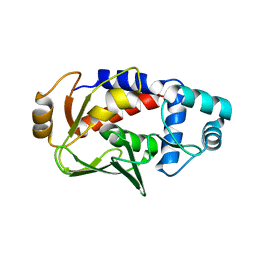 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | 分子名称: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | 著者 | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | 登録日 | 2008-10-25 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F0O
 
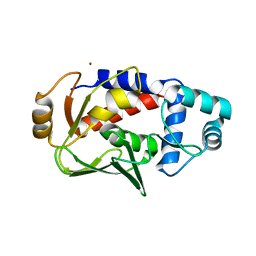 | | Crystal structure of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | 分子名称: | Alkylmercury lyase, BROMIDE ION | | 著者 | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | 登録日 | 2008-10-25 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3CMH
 
 | | SYNTHETIC LINEAR TRUNCATED ENDOTHELIN-1 AGONIST | | 分子名称: | PROTEIN (ENDOTHELIN-1) | | 著者 | Hewage, C.M, Jiang, L, Parkinson, J.A, Ramage, R, Sadler, I.H. | | 登録日 | 1998-09-03 | | 公開日 | 1999-09-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a novel ETB receptor selective agonist ET1-21 [Cys(Acm)1,15, Aib3,11, Leu7] by nuclear magnetic resonance spectroscopy and molecular modelling.
J.Pept.Res., 53, 1999
|
|
1VD7
 
 | | Solution structure of FMBP-1 tandem repeat 1 | | 分子名称: | Fibroin-modulator-binding-protein-1 | | 著者 | Kawaguchi, K, Yamaki, T, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | 登録日 | 2004-03-20 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of FMBP-1 tandem repeat 1
To be Published
|
|
1VD8
 
 | | Solution structure of FMBP-1 tandem repeat 2 | | 分子名称: | fibroin-modulator-binding-protein-1 | | 著者 | Yamaki, T, Kawaguchi, K, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | 登録日 | 2004-03-20 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of FMBP-1 tandem repeat 2
To be Published
|
|
1DNY
 
 | | SOLUTION STRUCTURE OF PCP, A PROTOTYPE FOR THE PEPTIDYL CARRIER DOMAINS OF MODULAR PEPTIDE SYNTHETASES | | 分子名称: | NON-RIBOSOMAL PEPTIDE SYNTHETASE PEPTIDYL CARRIER PROTEIN | | 著者 | Weber, T, Baumgartner, R, Renner, C, Marahiel, M.A, Holak, T.A. | | 登録日 | 1999-12-17 | | 公開日 | 2000-05-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases.
Structure Fold.Des., 8, 2000
|
|
1W3E
 
 | | Ribosomal L30e of Thermococcus celer, P59A mutant | | 分子名称: | 50S RIBOSOMAL PROTEIN L30E | | 著者 | Ma, H.W, Lee, C.F, Allen, M.D, Bycroft, M, Wong, K.B. | | 登録日 | 2004-07-15 | | 公開日 | 2006-10-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Role of Proline Residues in Thermostability of T. Celer L30E Protein
To be Published
|
|
7SGT
 
 | |
1F4S
 
 | | STRUCTURE OF TRANSCRIPTIONAL FACTOR ALCR IN COMPLEX WITH A TARGET DNA | | 分子名称: | DNA (5'-D(P*CP*GP*TP*GP*CP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*GP*CP*AP*CP*G)-3'), ETHANOL REGULON TRANSCRIPTIONAL FACTOR, ... | | 著者 | Cahuzac, B, Cerdan, R, Felenbok, B, Guittet, E. | | 登録日 | 2000-06-09 | | 公開日 | 2001-09-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein.
Structure, 9, 2001
|
|
5FRH
 
 | | Solution structure of oxidised RsrA | | 分子名称: | ANTI-SIGMA FACTOR RSRA | | 著者 | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | 登録日 | 2015-12-17 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
8BWW
 
 | |
7SXI
 
 | |
5U9X
 
 | | Ocellatin-LB2 | | 分子名称: | Ocellatin-K1 | | 著者 | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | 登録日 | 2016-12-18 | | 公開日 | 2017-03-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Ocellatin-LB2
To Be Published
|
|
6MY2
 
 | |
6MY3
 
 | |
2WK3
 
 | |
8BM7
 
 | |
