1B2Z
 
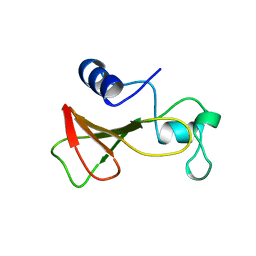 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | 分子名称: | PROTEIN (BARNASE), ZINC ION | | 著者 | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | 登録日 | 1998-12-03 | | 公開日 | 1998-12-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1AUG
 
 | | CRYSTAL STRUCTURE OF THE PYROGLUTAMYL PEPTIDASE I FROM BACILLUS AMYLOLIQUEFACIENS | | 分子名称: | PYROGLUTAMYL PEPTIDASE-1 | | 著者 | Odagaki, Y, Hayashi, A, Okada, K, Hirotsu, K, Kabashima, T, Ito, K, Yoshimoto, T, Tsuru, D, Sato, M, Clardy, J. | | 登録日 | 1997-08-26 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of pyroglutamyl peptidase I from Bacillus amyloliquefaciens reveals a new structure for a cysteine protease.
Structure Fold.Des., 7, 1999
|
|
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | 著者 | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | 登録日 | 1999-02-15 | | 公開日 | 1999-02-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
4JXS
 
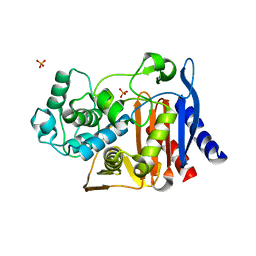 | |
1BAM
 
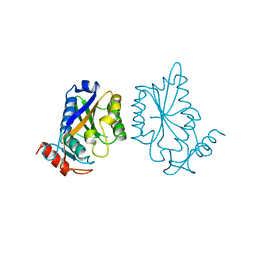 | |
1R6W
 
 | | Crystal structure of the K133R mutant of o-Succinylbenzoate synthase (OSBS) from Escherichia coli. Complex with SHCHC | | 分子名称: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, MAGNESIUM ION, o-Succinylbenzoate Synthase | | 著者 | Klenchin, V.A, Taylor Ringia, E.A, Gerlt, J.A, Rayment, I. | | 登録日 | 2003-10-17 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural and Mutagenic Studies of the Mechanism of the Reaction Catalyzed by o-Succinylbenzoate Synthase from Escherichia coli
Biochemistry, 42, 2003
|
|
1AY7
 
 | | RIBONUCLEASE SA COMPLEX WITH BARSTAR | | 分子名称: | BARSTAR, GUANYL-SPECIFIC RIBONUCLEASE SA | | 著者 | Sevcik, J, Urbanikova, L, Dauter, Z, Wilson, K.S. | | 登録日 | 1997-11-14 | | 公開日 | 1999-03-02 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Recognition of RNase Sa by the inhibitor barstar: structure of the complex at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1B20
 
 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | 分子名称: | PROTEIN (BARNASE), ZINC ION | | 著者 | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | 登録日 | 1998-12-03 | | 公開日 | 1998-12-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B3S
 
 | |
1B2S
 
 | |
4J8P
 
 | |
1B57
 
 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE IN COMPLEX WITH PHOSPHOGLYCOLOHYDROXAMATE | | 分子名称: | CHLORIDE ION, PHOSPHOGLYCOLOHYDROXAMIC ACID, PROTEIN (FRUCTOSE-BISPHOSPHATE ALDOLASE II), ... | | 著者 | Hall, D.R, Hunter, W.N. | | 登録日 | 1999-01-12 | | 公開日 | 2000-01-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity.
J.Mol.Biol., 287, 1999
|
|
7CFS
 
 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | 登録日 | 2020-06-28 | | 公開日 | 2020-10-21 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
1BSA
 
 | |
7COZ
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-41 | | 分子名称: | 1,2-ETHANEDIOL, 1-[3-(2-methyl-4-thiophen-2-yl-1,3-thiazol-5-yl)propanoyl]piperidine-4-carboxamide, Glutaminyl-peptide cyclotransferase, ... | | 著者 | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | 登録日 | 2020-08-05 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
1BNJ
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 9.0 | | 分子名称: | BARNASE | | 著者 | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | 登録日 | 1995-05-17 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1BRK
 
 | | BARNASE MUTANT WITH ILE 96 REPLACED BY ALA | | 分子名称: | BARNASE, ZINC ION | | 著者 | Cramer, P.C, Buckle, A, Fersht, A. | | 登録日 | 1995-03-09 | | 公開日 | 1995-07-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1BNE
 
 | |
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | 分子名称: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | 著者 | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | 登録日 | 2009-04-15 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
1TVR
 
 | | HIV-1 RT/9-CL TIBO | | 分子名称: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | 著者 | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | 登録日 | 1996-04-16 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
1CJC
 
 | |
1RHK
 
 | |
4GUI
 
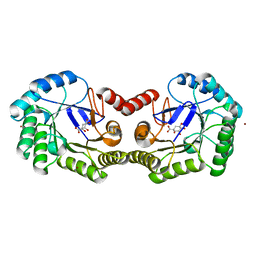 | | 1.78 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Quinate | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | 著者 | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-08-29 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
7BV8
 
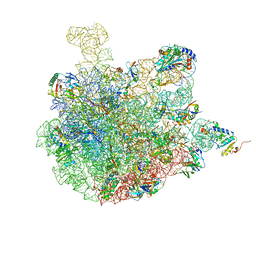 | | Mature 50S ribosomal subunit from RrmJ knock out E.coli strain | | 分子名称: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | 著者 | Wang, W, Li, W.Q, Ge, X.L, Yan, K.G, Mandava, C.S, Sanyal, S, Gao, N. | | 登録日 | 2020-04-09 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Loss of a single methylation in 23S rRNA delays 50S assembly at multiple late stages and impairs translation initiation and elongation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CFT
 
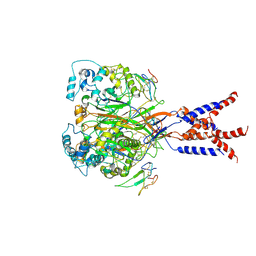 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | 著者 | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | 登録日 | 2020-06-28 | | 公開日 | 2020-10-21 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
