4UHG
 
 | |
4UI5
 
 | |
4UVN
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3-(4- chlorophenyl)-1,2-dihydroisoquinolin-1-one | | 分子名称: | 5-amino-3-(4-chlorophenyl)isoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | 著者 | Narwal, M, Haikarainen, T, Lehtio, L. | | 登録日 | 2014-08-07 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UUH
 
 | | X-ray crystal structure of human TNKS in complex with a small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 5-methyl-3-[4-(piperazin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, ... | | 著者 | Oliver, A.W, Rajasekaran, M.B, Pearl, L.H. | | 登録日 | 2014-07-28 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1- Ones as Potent and Selective Tankyrase Inhibitors
Medchemcommm, 6, 2015
|
|
4UVU
 
 | | Crystal structure of human tankyrase 2 in complex with 1-((4-(5- methyl-1-oxo-1,2-dihydroisoquinolin-3-yl)phenyl)methyl)pyrrolidin-1- ium | | 分子名称: | 5-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | 著者 | Haikarainen, T, Narwal, M, Lehtio, L. | | 登録日 | 2014-08-08 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVS
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- pentyl-1,2-dihydroisoquinolin-1-one | | 分子名称: | 5-amino-3-pentylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | 著者 | Narwal, M, Haikarainen, T, Lehtio, L. | | 登録日 | 2014-08-08 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
8HI9
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with Robinetin | | 分子名称: | 3,7-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, 3C-like proteinase nsp5 | | 著者 | Su, H.X, Xie, H, Li, M.J, Xu, Y.C. | | 登録日 | 2022-11-19 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Discovery of Polyphenolic Natural Products as SARS-CoV-2 M pro Inhibitors for COVID-19.
Pharmaceuticals, 16, 2023
|
|
4UX4
 
 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-(4- methylphenyl)-5-oxo-5,6-dihydro-1,6-naphthyridin-1-ium | | 分子名称: | (1S)-1-methyl-7-(4-methylphenyl)-5-oxo-1,5-dihydro-1,6-naphthyridin-1-ium, GLYCEROL, SULFATE ION, ... | | 著者 | Haikarainen, T, Lehtio, L. | | 登録日 | 2014-08-19 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Based Design, Synthesis and Evaluation in Vitro of Arylnaphthyridinones, Arylpyridopyrimidinones and Their Tetrahydro Derivatives as Inhibitors of the Tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
4D10
 
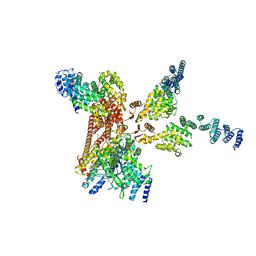 | | Crystal structure of the COP9 signalosome | | 分子名称: | COP9 SIGNALOSOME COMPLEX SUBUNIT 1, COP9 SIGNALOSOME COMPLEX SUBUNIT 2, COP9 SIGNALOSOME COMPLEX SUBUNIT 3, ... | | 著者 | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | 登録日 | 2014-04-30 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Crystal Structure of the Human Cop9 Signalosome
Nature, 512, 2014
|
|
4V29
 
 | |
8HGZ
 
 | | Crystal structure of insulin | | 分子名称: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | 著者 | DeMirci, H, Ayan, E. | | 登録日 | 2022-11-16 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Comparative Study of High-Resolution LysB29(N epsilon-myristoyl) des(B30) Insulin Structures Display Novel Dynamic Causal Interrelations in Monomeric-Dimeric Motions
Crystals, 13, 2023
|
|
8ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | 分子名称: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION | | 著者 | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | 登録日 | 1990-05-30 | | 公開日 | 1991-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
4XUR
 
 | |
4XUO
 
 | |
4XUN
 
 | |
6QAJ
 
 | | Structure of the tripartite motif of KAP1/TRIM28 | | 分子名称: | Endolysin,Transcription intermediary factor 1-beta, ZINC ION | | 著者 | Stoll, G.A, Oda, S, Yu, M, Modis, Y. | | 登録日 | 2018-12-19 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.901 Å) | | 主引用文献 | Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4BKW
 
 | | Crystal structure of the C-terminal region of human ZFYVE9 | | 分子名称: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, ZINC FINGER FYVE DOMAIN-CONTAINING PROTEIN 9 | | 著者 | Williams, E, Krojer, T, Vollmar, M, Shrestha, L, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | 登録日 | 2013-04-30 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Crystal Structure of the C-Terminal Region of Human Zfyve9
Ph D Thesis, 2013
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8JNQ
 
 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with a substrate compound c | | 分子名称: | (1Z,3E,5E,7S,8R,10S,11R,13R,15R,16E,18E,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.09,13.08,15]octacosa-1(2),3,5,16,18-pentaene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Jiang, P, Zhang, L.P, Zhang, C.S. | | 登録日 | 2023-06-06 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNP
 
 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with the substrate ikarugamycin | | 分子名称: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450 CftA, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Jiang, P, Zhang, L.P, Zhang, C.S. | | 登録日 | 2023-06-06 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4AZZ
 
 | |
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
