6HEQ
 
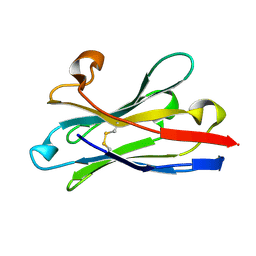 | | Prion nanobody 484 | | 分子名称: | Prion nanobody 484 | | 著者 | Soror, S.H, Abskharon, R.N, Wohlkonig, A. | | 登録日 | 2018-08-20 | | 公開日 | 2019-12-04 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
5JQH
 
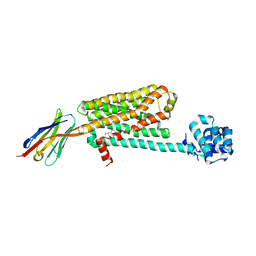 | | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 | | 分子名称: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, Endolysin,Beta-2 adrenergic receptor, ... | | 著者 | Staus, D.P, Strachan, R.T, Manglik, A, Pani, B, Kahsai, A.W, Kim, T.H, Wingler, L.M, Ahn, S, Chatterjee, A, Masoudi, A, Kruse, A.C, Pardon, E, Steyaert, J, Weis, W.I, Prosser, R.S, Kobilka, B.K, Costa, T, Lefkowitz, R.J. | | 登録日 | 2016-05-05 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation.
Nature, 535, 2016
|
|
5K79
 
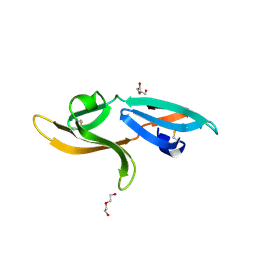 | | Structure and anti-HIV activity of CYT-CVNH, a new cyanovirin-n homolog | | 分子名称: | 1,2-ETHANEDIOL, Cyanovirin-N domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Matei, E, Basu, R, Furey, W, Shi, J, Calnan, C, Aiken, C, Gronenborn, A.M. | | 登録日 | 2016-05-25 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and Glycan Binding of a New Cyanovirin-N Homolog.
J.Biol.Chem., 291, 2016
|
|
3JBG
 
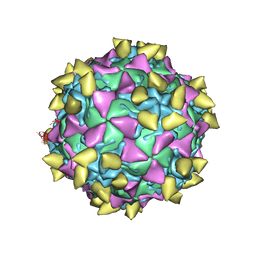 | | Complex of poliovirus with VHH PVSS21E | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | 登録日 | 2015-08-26 | | 公開日 | 2016-01-27 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
5XYA
 
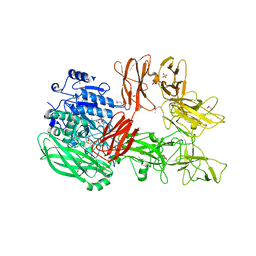 | | Crystal structure of a serine protease from Streptococcus species | | 分子名称: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, Chemokine protease C, ... | | 著者 | Jobichen, C, Sivaraman, J. | | 登録日 | 2017-07-06 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
5GNB
 
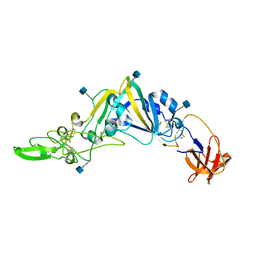 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | 登録日 | 2016-07-20 | | 公開日 | 2017-06-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
1E09
 
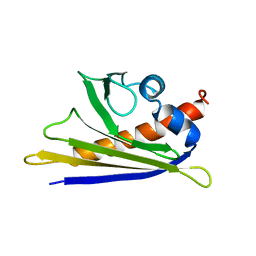 | | Solution Structure of the Major Cherry Allergen Pru av 1 | | 分子名称: | PRU AV 1 | | 著者 | Neudecker, P, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | 登録日 | 2000-03-15 | | 公開日 | 2001-03-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Allergic Cross-Reactivity Made Visible: The Solution Structure of the Major Cherry Allergen Pru Av 1
J.Biol.Chem., 276, 2001
|
|
5KA5
 
 | |
1GWV
 
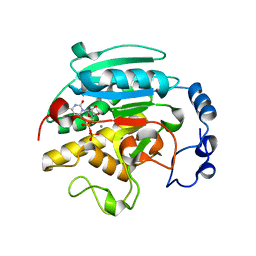 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - LACTOSE COMPLEX | | 分子名称: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | 登録日 | 2002-03-26 | | 公開日 | 2003-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1H4B
 
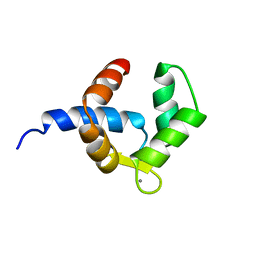 | | SOLUTION STRUCTURE OF THE BIRCH POLLEN ALLERGEN BET V 4 | | 分子名称: | CALCIUM ION, POLCALCIN BET V 4 | | 著者 | Neudecker, P, Nerkamp, J, Eisenmann, A, Lauber, T, Lehmann, K, Schweimer, K, Roesch, P. | | 登録日 | 2003-02-26 | | 公開日 | 2004-02-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure, Dynamics, and Hydrodynamics of the Calcium-Bound Cross-Reactive Birch Pollen Allergen Bet V 4 Reveal a Canonical Monomeric Two EF-Hand Assembly with a Regulatory Function
J.Mol.Biol., 336, 2004
|
|
1Q8M
 
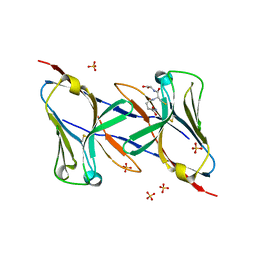 | | Crystal structure of the human myeloid cell activating receptor TREM-1 | | 分子名称: | GLUTATHIONE, SULFATE ION, triggering receptor expressed on myeloid cells 1 | | 著者 | Radaev, S, Kattah, M, Rostro, B, Colonna, M, Sun, P.D. | | 登録日 | 2003-08-21 | | 公開日 | 2003-12-09 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the human myeloid cell activating receptor TREM-1
Structure, 11, 2003
|
|
1GWW
 
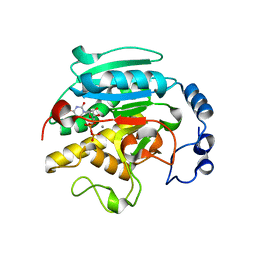 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - ALPHA-D-GLUCOSE COMPLEX | | 分子名称: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | 登録日 | 2002-03-26 | | 公開日 | 2003-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
5XYR
 
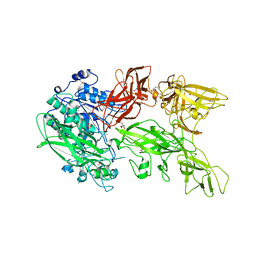 | | Crystal structure of a serine protease from Streptococcus species | | 分子名称: | CALCIUM ION, CHLORIDE ION, Chemokine protease C, ... | | 著者 | Jobichen, C, Sivaraman, J. | | 登録日 | 2017-07-10 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
3Q1J
 
 | | Crystal structure of tudor domain 1 of human PHD finger protein 20 | | 分子名称: | PHD finger protein 20, UNKNOWN ATOM OR ION | | 著者 | Tempel, W, Li, Z, Wernimont, A.K, Chao, X, Bian, C, Lam, R, Crombet, L, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-12-17 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins.
Febs Lett., 586, 2012
|
|
1NA8
 
 | | Crystal structure of ADP-ribosylation factor binding protein GGA1 | | 分子名称: | ADP-ribosylation factor binding protein GGA1 | | 著者 | Lui, W.W, Collins, B.M, Hirst, J, Motley, A, Millar, C, Schu, P, Owen, D.J, Robinson, M.S. | | 登録日 | 2002-11-27 | | 公開日 | 2003-07-29 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin
Mol.Cell.Biol., 14, 2003
|
|
3ULA
 
 | | Crystal structure of the TV3 mutant F63W-MD-2-Eritoran complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-O-DECYL-2-DEOXY-6-O-{2-DEOXY-3-O-[(3R)-3-METHOXYDECYL]-6-O-METHYL-2-[(11Z)-OCTADEC-11-ENOYLAMINO]-4-O-PHOSPHONO-BETA-D-GLUCOPYRANOSYL}-2-[(3-OXOTETRADECANOYL)AMINO]-1-O-PHOSPHONO-ALPHA-D-GLUCOPYRANOSE, Lymphocyte antigen 96, ... | | 著者 | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | 登録日 | 2011-11-10 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
6IM7
 
 | | CueO-12.1 multicopper oxidase | | 分子名称: | Blue copper oxidase CueO,12.1 peptide,Blue copper oxidase CueO, CALCIUM ION | | 著者 | Wongsantichon, J, Robinson, R, Ghadessy, F. | | 登録日 | 2018-10-22 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
1GX4
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - N-ACETYL LACTOSAMINE COMPLEX | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | 著者 | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | 登録日 | 2002-03-27 | | 公開日 | 2003-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
5YGI
 
 | |
3KXS
 
 | |
1O7O
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | 分子名称: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | 登録日 | 2002-11-11 | | 公開日 | 2003-11-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity.
Biochemistry, 42, 2003
|
|
1SMO
 
 | | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47 . | | 分子名称: | L(+)-TARTARIC ACID, triggering receptor expressed on myeloid cells 1 | | 著者 | Kelker, M.S, Foss, T.R, Peti, W, Teyton, L, Kelly, J.W, Wilson, I.A. | | 登録日 | 2004-03-09 | | 公開日 | 2004-09-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47A.
J.Mol.Biol., 342, 2004
|
|
4WUA
 
 | | Crystal structure of human SRPK1 complexed to an inhibitor SRPIN340 | | 分子名称: | CITRIC ACID, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]-4-pyridinecarboxamide, SRSF protein kinase 1, ... | | 著者 | Hoshina, M, Ikura, T, Hosoya, T, Hagiwara, M, Ito, N. | | 登録日 | 2014-10-31 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of a Dual Inhibitor of SRPK1 and CK2 That Attenuates Pathological Angiogenesis of Macular Degeneration in Mice
Mol.Pharmacol., 88, 2015
|
|
8OKL
 
 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKK
 
 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
