7NL0
 
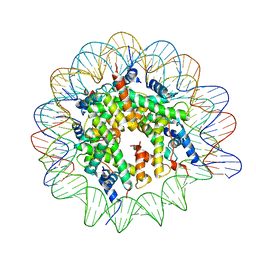 | | Cryo-EM structure of the Lin28B nucleosome core particle | | 分子名称: | DNA (131-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Roberts, G.A, Ozkan, B, Gachulincova, I, O Dwyer, M.R, Hall-Ponsele, E, Saxena, M, Robinson, P.J, Soufi, A. | | 登録日 | 2021-02-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Dissecting OCT4 defines the role of nucleosome binding in pluripotency.
Nat.Cell Biol., 23, 2021
|
|
7WBX
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2021-12-17 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBW
 
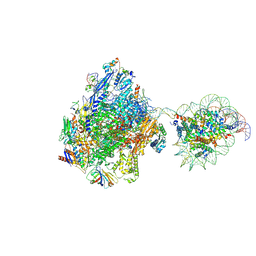 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2021-12-17 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBV
 
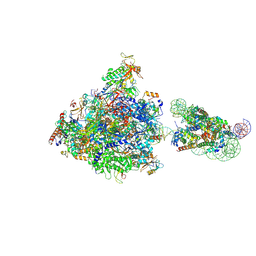 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome | | 分子名称: | DNA (159-MER), DNA (198-MER), DNA-directed RNA polymerase subunit, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2021-12-17 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7TOR
 
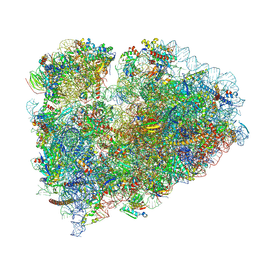 | | Mammalian 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein GR20 | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | 登録日 | 2022-01-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
5JA4
 
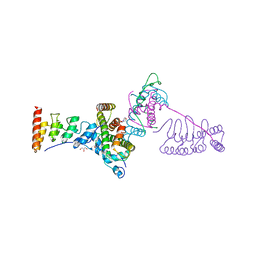 | |
5KDM
 
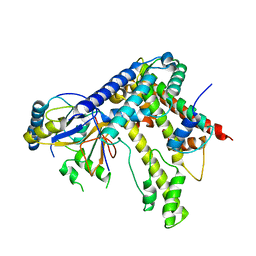 | |
5KGF
 
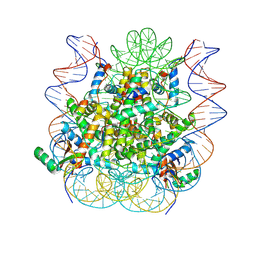 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | 分子名称: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | 著者 | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | 登録日 | 2016-06-13 | | 公開日 | 2016-07-27 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.54 Å) | | 主引用文献 | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
7V1M
 
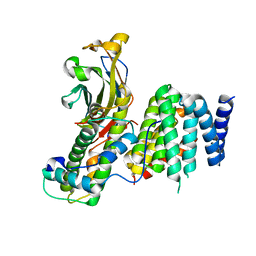 | |
7UCJ
 
 | | Mammalian 80S translation initiation complex with mRNA and Harringtonine | | 分子名称: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | 著者 | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | 登録日 | 2022-03-16 | | 公開日 | 2022-06-01 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
7U51
 
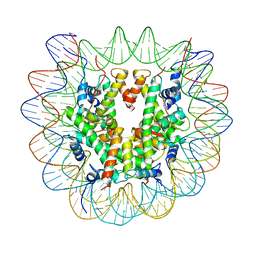 | |
7U50
 
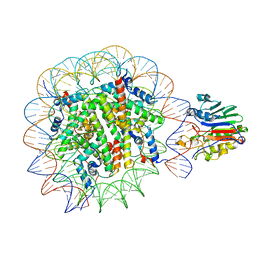 | |
7U52
 
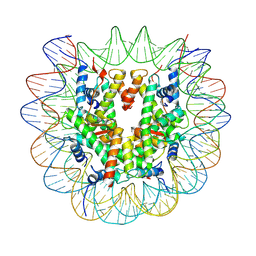 | |
7N9H
 
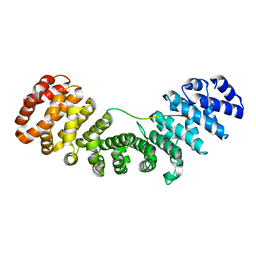 | |
7U53
 
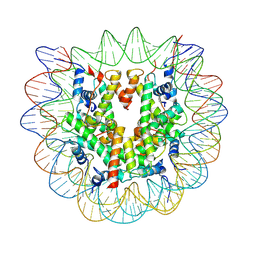 | |
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
5IFM
 
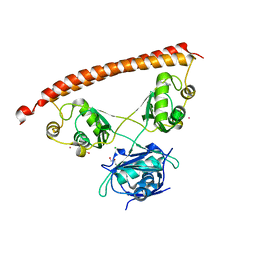 | | Human NONO (p54nrb) Homodimer | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-POU domain-containing octamer-binding protein, ... | | 著者 | Knott, G.J, Bond, C.S. | | 登録日 | 2016-02-26 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7O7Z
 
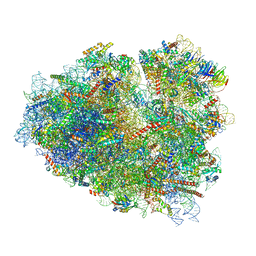 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7UPZ
 
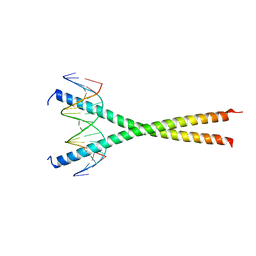 | | Structural basis for cell type specific DNA binding of C/EBPbeta: the case of cell cycle inhibitor p15INK4b promoter | | 分子名称: | CCAAT/enhancer-binding protein beta, DNA (5'-D(*AP*TP*TP*CP*TP*TP*AP*AP*GP*AP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*CP*TP*TP*TP*CP*TP*TP*AP*AP*GP*AP*A)-3') | | 著者 | Lountos, G.T, Cherry, S, Tropea, J.E, Wlodawer, A, Miller, M. | | 登録日 | 2022-04-18 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.487 Å) | | 主引用文献 | Structural basis for cell type specific DNA binding of C/EBP beta : The case of cell cycle inhibitor p15INK4b promoter.
J.Struct.Biol., 214, 2022
|
|
7OBR
 
 | | RNC-SRP early complex | | 分子名称: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Jomaa, A, Ban, N. | | 登録日 | 2021-04-23 | | 公開日 | 2021-07-21 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
7W9V
 
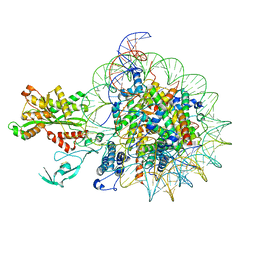 | | Cryo-EM structure of nucleosome in complex with p300 acetyltransferase catalytic core (complex I) | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Hatazawa, S, Liu, J, Takizawa, Y, Zandian, M, Negishi, L, Kutateladze, T.G, Kurumizaka, H. | | 登録日 | 2021-12-10 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Structural basis for binding diversity of acetyltransferase p300 to the nucleosome.
Iscience, 25, 2022
|
|
7M3X
 
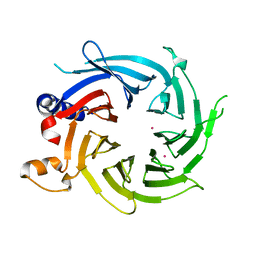 | | Crystal Structure of the Apo Form of Human RBBP7 | | 分子名称: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | 著者 | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2021-03-19 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
7MQA
 
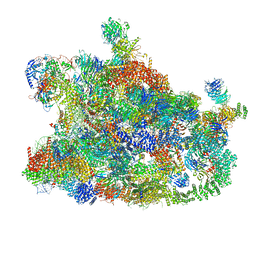 | | Cryo-EM structure of the human SSU processome, state post-A1 | | 分子名称: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | 著者 | Vanden Broeck, A, Singh, S, Klinge, S. | | 登録日 | 2021-05-05 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
