8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.68 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
7Z9C
 
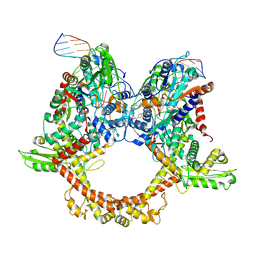 | | E.coli gyrase holocomplex with 217 bp DNA and albicidin | | 分子名称: | DNA (5'-D(*AP*AP*TP*CP*AP*CP*CP*CP*GP*CP*AP*CP*AP*GP*AP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*TP*TP*AP*TP*GP*CP*CP*TP*GP*AP*TP*TP*CP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*CP*TP*GP*TP*GP*CP*GP*GP*GP*T)-3'), ... | | 著者 | Ghilarov, D, Heddle, J.G.H, Suessmuth, R. | | 登録日 | 2022-03-21 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Nat Catal, 6, 2023
|
|
8WH4
 
 | |
6CEU
 
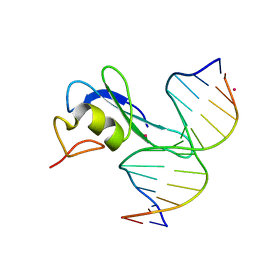 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | 著者 | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-12 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
7RHZ
 
 | |
8FDP
 
 | |
5D50
 
 | | Crystal structure of Rep-Ant complex from Salmonella-temperate phage | | 分子名称: | Anti-repressor protein, Repressor | | 著者 | Son, S.H, Yoon, H.J, Ryu, S, Lee, H.H. | | 登録日 | 2015-08-10 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4Z
 
 | |
6OUN
 
 | |
6OTZ
 
 | |
5FD3
 
 | | Structure of Lin54 tesmin domain bound to DNA | | 分子名称: | DNA (5'-D(*CP*AP*GP*TP*TP*TP*CP*AP*AP*AP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*TP*TP*TP*GP*AP*AP*AP*CP*T)-3'), Protein lin-54 homolog, ... | | 著者 | Marceau, A.H, Felthousen, J.G, Goetsch, P.D, Lee, H, Tripathi, S.M, Strome, S, Litovchick, L, Rubin, S.M. | | 登録日 | 2015-12-15 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters.
Nat Commun, 7, 2016
|
|
3KLF
 
 | | Crystal structure of wild-type HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | 分子名称: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), GLYCEROL, ... | | 著者 | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | 登録日 | 2009-11-07 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KLE
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | 分子名称: | DNA (25-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), GLYCEROL, ... | | 著者 | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | 登録日 | 2009-11-07 | | 公開日 | 2010-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8DVP
 
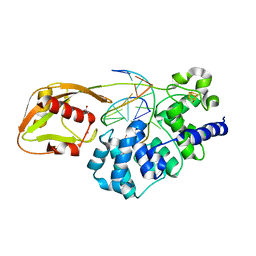 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with substrate purine | | 分子名称: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | 著者 | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | 登録日 | 2022-07-29 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DW0
 
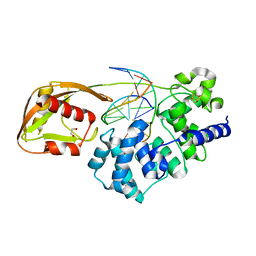 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an enzyme-generated abasic site (AP) product and crystallized with sodium acetate | | 分子名称: | 1,2-ETHANEDIOL, Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | 著者 | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | 登録日 | 2022-07-30 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DW4
 
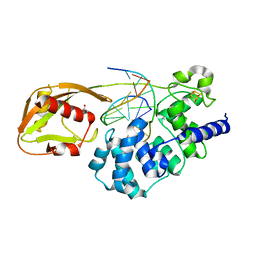 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an abasic site product (AP) generated by the enzyme in crystals by removal of calcium | | 分子名称: | ACETATE ION, Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | 著者 | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | 登録日 | 2022-07-31 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
5KBJ
 
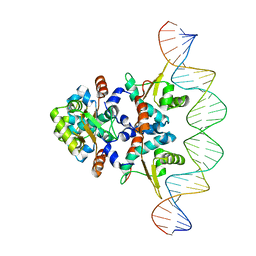 | | Structure of Rep-DNA complex | | 分子名称: | DNA (32-MER), Replication initiator A, N-terminal | | 著者 | Schumacher, M. | | 登録日 | 2016-06-03 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
8XPM
 
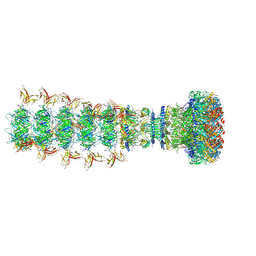 | |
5Z51
 
 | |
7KZG
 
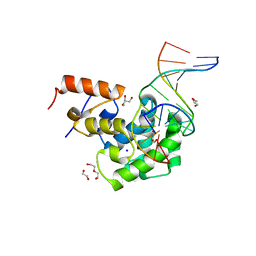 | | Human MBD4 glycosylase domain bound to DNA containing oxacarbenium-ion analog 1-aza-2'-deoxyribose | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | 登録日 | 2020-12-10 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
7KZ1
 
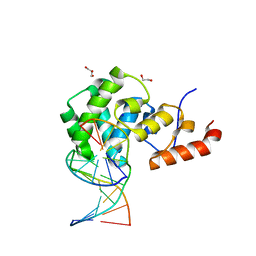 | | Human MBD4 glycosylase domain bound to DNA containing an abasic site | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*CP*GP*(ORP)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), ... | | 著者 | Pidugu, L.S, Bright, H, Pozharski, E, Drohat, A.C. | | 登録日 | 2020-12-09 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
1D9F
 
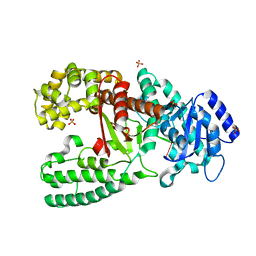 | | CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA TETRAMER CARRYING 2'-O-(3-AMINOPROPYL)-RNA MODIFICATION 5'-D(TT)-AP(U)-D(T)-3' | | 分子名称: | DNA POLYMERASE I, DNA/RNA (5'-D(*TP*TP)-R(*(U31)P)-D(*T)-3'), SULFATE ION, ... | | 著者 | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | 登録日 | 1999-10-27 | | 公開日 | 1999-12-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural origins of the exonuclease resistance of a zwitterionic RNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6X9I
 
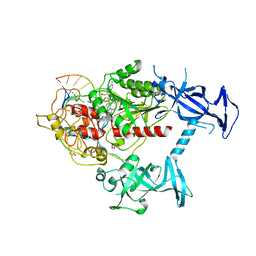 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | 著者 | Pathuri, S, Horton, J.R, Cheng, X. | | 登録日 | 2020-06-02 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
7MVS
 
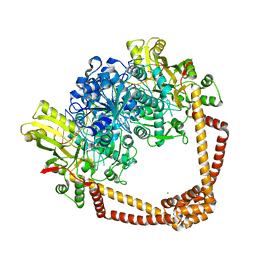 | | DNA gyrase complexed with uncleaved DNA and Compound 7 to 2.6A resolution | | 分子名称: | (1S)-2-[(2r,5S)-5-{[(2,3-dihydro-1,4-benzodioxin-6-yl)methyl]amino}-1,3-dioxan-2-yl]-1-(3-fluoro-6-methoxyquinolin-4-yl)ethan-1-ol, CHLORIDE ION, DNA (5'-D(P*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Ratigan, S.C, McElroy, C.A. | | 登録日 | 2021-05-15 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.60137153 Å) | | 主引用文献 | Optimization of TopoIV Potency, ADMET Properties, and hERG Inhibition of 5-Amino-1,3-dioxane-Linked Novel Bacterial Topoisomerase Inhibitors: Identification of a Lead with In Vivo Efficacy against MRSA.
J.Med.Chem., 64, 2021
|
|
