1ODO
 
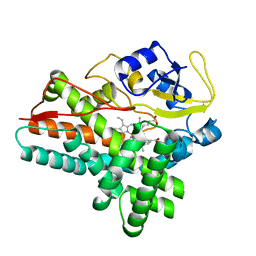 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | 分子名称: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | 著者 | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | 登録日 | 2003-02-19 | | 公開日 | 2004-01-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
8ETN
 
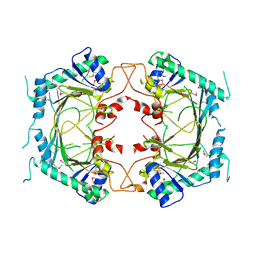 | |
5OCJ
 
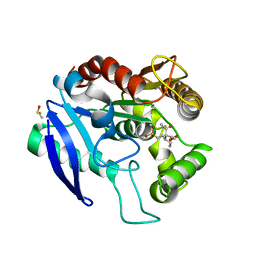 | | Crystal structure of Ag85C bound to cyclophostin 8beta inhibitor | | 分子名称: | DIMETHYL SULFOXIDE, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, methoxy-[(3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]pentadecyl]phosphinic acid | | 著者 | Viljoen, A, Richard, M, Nguyen, P.C, Spilling, C.D, Canaan, S, Cavalier, J.F, Blaise, M, Kremer, L. | | 登録日 | 2017-07-03 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cyclipostins and cyclophostin analogs inhibit the antigen 85C from
J. Biol. Chem., 293, 2018
|
|
5OVL
 
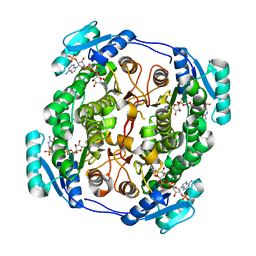 | | crystal structure of MabA bound to NADP+ from M. smegmatis | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | 登録日 | 2017-08-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5O15
 
 | | Crystal structure of bifunctional dehydratase-cyclase domain in ambruticin biosynthesis | | 分子名称: | AmbC, GLYCEROL | | 著者 | Sung, K.H, Berkhan, G, Hollmann, T, Wagner, L, Hahn, F, Blankenfeldt, W. | | 登録日 | 2017-05-18 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.174 Å) | | 主引用文献 | Insights into the Dual Activity of a Bifunctional Dehydratase-Cyclase Domain.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8CIE
 
 | | Crystal structure of the human CDKL5 kinase domain with compound YL-354 | | 分子名称: | 4-[[3,5-bis(fluoranyl)phenyl]carbonylamino]-~{N}-piperidin-4-yl-1~{H}-pyrazole-3-carboxamide, Cyclin-dependent kinase-like 5, SULFATE ION | | 著者 | Richardson, W, Chen, X, Newman, J.A, Bakshi, S, Lakshminarayana, B, Brooke, L, Bullock, A.N. | | 登録日 | 2023-02-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective.
Acs Chem Neurosci, 14, 2023
|
|
5L02
 
 | | S324T variant of B. pseudomallei KatG | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Catalase-peroxidase, PHOSPHATE ION, ... | | 著者 | Loewen, P.C. | | 登録日 | 2016-07-26 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural characterization of the Ser324Thr variant of the catalase-peroxidase (KatG) from Burkholderia pseudomallei
J. Mol. Biol., 345, 2005
|
|
6WJ9
 
 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | 分子名称: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | 登録日 | 2020-04-13 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6WJB
 
 | | UDP-GlcNAc C4-epimerase from Pseudomonas protegens in complex with NAD and UDP-GlcNAc | | 分子名称: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Marmont, L.S, Pfoh, R, Robinson, H, Howell, P.L. | | 登録日 | 2020-04-13 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6WJA
 
 | |
6XOU
 
 | |
6XOV
 
 | |
6XOS
 
 | |
6XOT
 
 | |
6J2U
 
 | |
7W43
 
 | |
6U1V
 
 | | Crystal structure of acyl-ACP/acyl-CoA dehydrogenase from allylmalonyl-CoA and FK506 biosynthesis, TcsD | | 分子名称: | Acyl-CoA dehydrogenase domain-containing protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | 著者 | Blake-Hedges, J.M, Pereira, J.H, Barajas, J.F, Adams, P.D, Keasling, J.D. | | 登録日 | 2019-08-16 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Mechanism of Regioselectivity in an Unusual Bacterial Acyl-CoA Dehydrogenase.
J.Am.Chem.Soc., 142, 2020
|
|
6ERK
 
 | | Crystal structure of diaminopelargonic acid aminotransferase from Psychrobacter cryohalolentis | | 分子名称: | 1,2-ETHANEDIOL, Aminotransferase, GLYCEROL, ... | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Bezsudnova, E.Y, Stekhanova, T.N, Rakitina, T.V, Popov, V.O. | | 登録日 | 2017-10-18 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Diaminopelargonic acid transaminase from Psychrobacter cryohalolentis is active towards (S)-(-)-1-phenylethylamine, aldehydes and alpha-diketones.
Appl. Microbiol. Biotechnol., 102, 2018
|
|
6ETO
 
 | |
6SK1
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with coenzyme A | | 分子名称: | ACETATE ION, COENZYME A, L-2,4-diaminobutyric acid acetyltransferase | | 著者 | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | 登録日 | 2019-08-14 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6VKZ
 
 | |
6VL1
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with NGG and Mg2+ | | 分子名称: | DabA, MAGNESIUM ION, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-L-glutamic acid | | 著者 | Chekan, J.R, Noel, J.P, Moore, B.S. | | 登録日 | 2020-01-22 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VL0
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with GSPP and Mn2+ | | 分子名称: | DabA, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Chekan, J.R, Noel, J.P, Moore, B.S. | | 登録日 | 2020-01-22 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6A9B
 
 | |
7KL8
 
 | | Structure of F420 binding protein Rv1558 from Mycobacterium tuberculosis with F420 bound | | 分子名称: | COENZYME F420, COENZYME F420-3, Deazaflavin-dependent nitroreductase, ... | | 著者 | Lee, B.M, Tan, L.L, Jackson, C.J. | | 登録日 | 2020-10-29 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.469 Å) | | 主引用文献 | Potency boost of a Mycobacterium tuberculosis dihydrofolate reductase inhibitor by multienzyme F 420 H 2 -dependent reduction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
