4G31
 
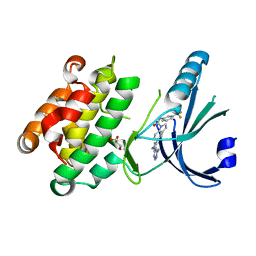 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | 分子名称: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | 著者 | Gampe, R.T, Axten, J.M. | | 登録日 | 2012-07-13 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
2HOS
 
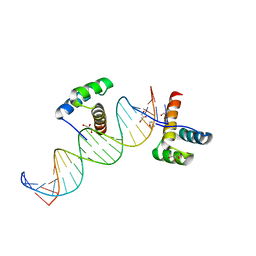 | | Phage-Selected Homeodomain Bound to Unmodified DNA | | 分子名称: | 3-METHYL-1,3-OXAZOLIDIN-2-ONE, 5'-D(*AP*TP*CP*CP*GP*GP*GP*GP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*CP*CP*CP*CP*GP*GP*A)-3', ... | | 著者 | Shokat, K.M, Feldman, M.E, Simon, M.D. | | 登録日 | 2006-07-16 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and properties of a re-engineered homeodomain protein-DNA interface.
Acs Chem.Biol., 1, 2006
|
|
4GFS
 
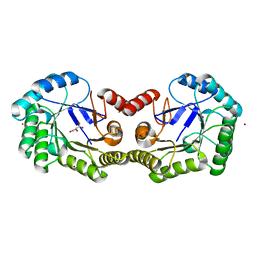 | | 1.8 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Nickel Bound at Active Site | | 分子名称: | 3-dehydroquinate dehydratase, NICKEL (II) ION, SUCCINIC ACID | | 著者 | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-08-03 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 1.8 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Nickel Bound at Active Site
TO BE PUBLISHED
|
|
3UUW
 
 | | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile. | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-11-28 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.
TO BE PUBLISHED
|
|
4KRZ
 
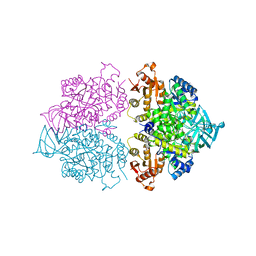 | | Apo crystal structure of pyruvate kinase (PYK) from Trypanosoma cruzi | | 分子名称: | GLYCEROL, POTASSIUM ION, Pyruvate kinase | | 著者 | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | 登録日 | 2013-05-17 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of pyruvate kinases display evolutionarily divergent allosteric strategies.
R Soc Open Sci, 1, 2014
|
|
1EMD
 
 | |
2GF3
 
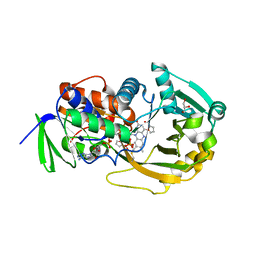 | | Structure of the complex of monomeric sarcosine with its substrate analogue inhibitor 2-furoic acid at 1.3 A resolution. | | 分子名称: | 2-FUROIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Chen, Z, Trickey, P, Jorns, M.S, Mathews, F.S. | | 登録日 | 2006-03-21 | | 公開日 | 2007-02-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure of the complex of monomeric sarcosine with its substrate analogue inhibitor 2-furoic acid at 1.3 A resolution.
TO BE PUBLISHED
|
|
4ESK
 
 | |
4E4V
 
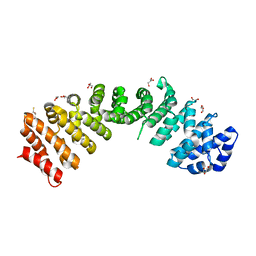 | | The crystal structure of the dimeric human importin alpha 1 at 2.5 angstrom resolution. | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Importin subunit alpha-2 | | 著者 | Hang, P.C, Miknis, Z.M, Franke, W.A, Umland, T.C, Schultz, L.W. | | 登録日 | 2012-03-13 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5283 Å) | | 主引用文献 | The crystal structure of the dimeric human importin alpha 1 at 2.5 angstrom resolution.
To be Published
|
|
7DBN
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2020-10-21 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
1WNB
 
 | | Escherichia coli YdcW gene product is a medium-chain aldehyde dehydrogenase (complexed with nadh and betaine aldehyde) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETAINE ALDEHYDE, Putative betaine aldehyde dehydrogenase | | 著者 | Gruez, A, Roig-Zamboni, V, Tegoni, M, Cambillau, C. | | 登録日 | 2004-07-29 | | 公開日 | 2004-10-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure and Kinetics Identify Escherichia coli YdcW Gene Product as a Medium-chain Aldehyde Dehydrogenase
J.Mol.Biol., 343, 2004
|
|
5DYW
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-((1-benzylpiperidin-3-yl)methyl)-N-(2-methoxyethyl)naphthalene-2-sulfonamide | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Coquelle, N, Brus, B, Colletier, J.P. | | 登録日 | 2015-09-25 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Development of an in-vivo active reversible butyrylcholinesterase inhibitor.
Sci Rep, 6, 2016
|
|
4DDM
 
 | | Pantothenate synthetase in complex with 2,1,3-benzothiadiazole-5-carboxylic acid | | 分子名称: | 1,2-ETHANEDIOL, 2,1,3-benzothiadiazole-5-carboxylic acid, ETHANOL, ... | | 著者 | Silvestre, H.L. | | 登録日 | 2012-01-18 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1X0V
 
 | |
1BXS
 
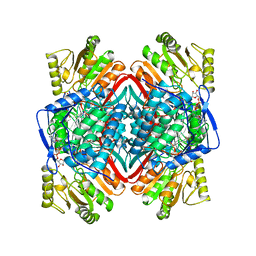 | | SHEEP LIVER CLASS 1 ALDEHYDE DEHYDROGENASE WITH NAD BOUND | | 分子名称: | ALDEHYDE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Moore, S.A, Baker, H.M, Blythe, T.J, Kitson, K.E, Kitson, T.M, Baker, E.N. | | 登録日 | 1998-10-08 | | 公開日 | 1999-04-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases.
Structure, 6, 1998
|
|
1GOF
 
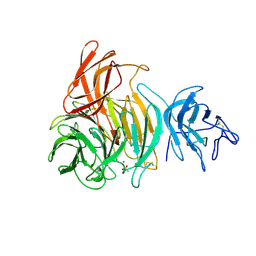 | | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE | | 分子名称: | ACETIC ACID, COPPER (II) ION, GALACTOSE OXIDASE, ... | | 著者 | Ito, N, Phillips, S.E.V, Knowles, P.F. | | 登録日 | 1993-09-30 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase.
Nature, 350, 1991
|
|
1BMD
 
 | |
3P4T
 
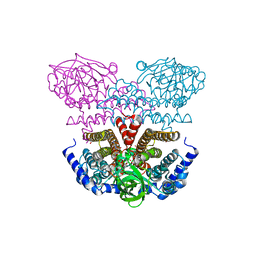 | | Crystal structure of a putative acyl-CoA dehydrogenase from Mycobacterium smegmatis | | 分子名称: | 1,2-ETHANEDIOL, Putative acyl-CoA dehydrogenase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(4aS,10aR)-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | 著者 | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2010-10-07 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Increasing the structural coverage of tuberculosis drug targets.
Tuberculosis (Edinb), 95, 2015
|
|
1GOG
 
 | |
3RU6
 
 | | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168 | | 分子名称: | CHLORIDE ION, IODIDE ION, Orotidine 5'-phosphate decarboxylase | | 著者 | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-05-04 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
1X0X
 
 | |
3OZ2
 
 | |
5FEC
 
 | |
3RMI
 
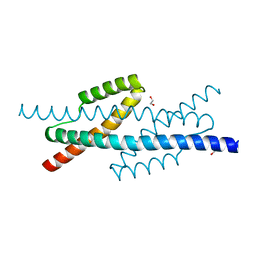 | |
5UC6
 
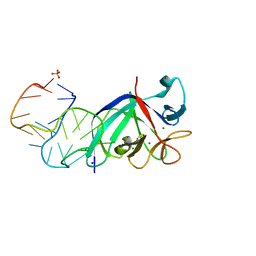 | |
