6TTG
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with LMD62 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-(2-morpholin-4-ylethoxy)-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | 著者 | Welin, M, Kimbung, R, Focht, D. | | 登録日 | 2019-12-27 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens.
Eur.J.Med.Chem., 213, 2021
|
|
3I6U
 
 | |
3I6W
 
 | |
2OBP
 
 | |
6RML
 
 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a 53BP1 phosphopeptide | | 分子名称: | 53BP1, DNA topoisomerase 2-binding protein 1 | | 著者 | Day, M, Oliver, A.W, Pearl, L.H. | | 登録日 | 2019-05-07 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint.
Elife, 8, 2019
|
|
5G34
 
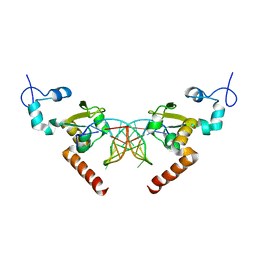 | | Structure of Rad14 in complex with acetylaminoanthracene-C8-guanine containing DNA | | 分子名称: | 5'-D(*GP*CP*TP*CP*TP*AP*6FKP*TP*CP*AP*TP*CP*AP*CP)-3', 5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*GP)-3', RAD14, ... | | 著者 | Simon, N, Ebert, C, Schneider, S. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Bulky Adduct DNA Lesion Recognition by the Nucleotide Excision Repair Protein Rad14.
Chemistry, 22, 2016
|
|
3OOL
 
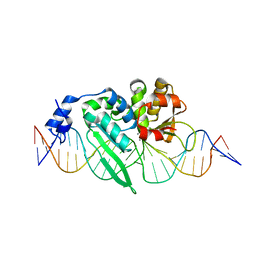 | | I-SceI complexed with C/G+4 DNA substrate | | 分子名称: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | 著者 | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | 登録日 | 2010-08-31 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
4ABT
 
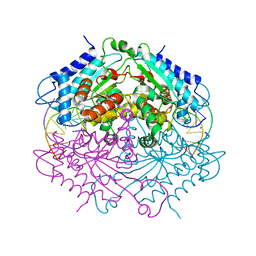 | | Crystal structure of Type IIF restriction endonuclease NgoMIV with cognate uncleaved DNA | | 分子名称: | 5'-D(*TP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP)-3', CALCIUM ION, TYPE-2 RESTRICTION ENZYME NGOMIV | | 著者 | Manakova, E.N, Grazulis, S, Zaremba, M, Tamulaitiene, G, Golovenko, D, Siksnys, V. | | 登録日 | 2011-12-11 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structure of Type Iif Restriction Endonuclease Ngomiv with Cognate Uncleaved DNA
To be Published
|
|
6RMM
 
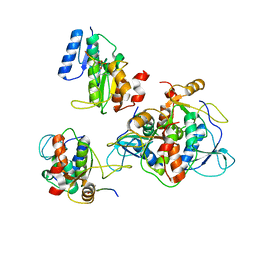 | |
8ALQ
 
 | |
8IF5
 
 | | AFB1-AF26 APTAMER COMPLEX | | 分子名称: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | 著者 | Xu, G.H, Wang, C, Li, C.G. | | 登録日 | 2023-02-17 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
7QWV
 
 | | Crystal structure of the REC114-TOPOVIBL complex. | | 分子名称: | Meiotic recombination protein REC114, Type 2 DNA topoisomerase 6 subunit B-like | | 著者 | Juarez-Martinez, A.B, Robert, T, de Massy, B, Kadlec, J. | | 登録日 | 2022-01-25 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks.
Nat Commun, 13, 2022
|
|
2M8G
 
 | |
5G35
 
 | | Structure of Rad14 in complex with acetylaminopyren-C8-guanine containing DNA | | 分子名称: | 5'-D(*GP*CP*TP*CP*TP*AP*8PYP*TP*CP*AP*TP*CP*AP*CP)-3', 5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*GP)-3', RAD14, ... | | 著者 | Simon, N, Ebert, C, Schneider, S. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Bulky Adduct DNA Lesion Recognition by the Nucleotide Excision Repair Protein Rad14.
Chemistry, 22, 2016
|
|
5G33
 
 | | Structure of Rad14 in complex with acetylnaphtyl-guanine containing DNA | | 分子名称: | 5'-D(*GP*CP*TP*CP*TP*AP*MFOP*TP*CP*AP*TP*CP*AP*CP)-3', 5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*GP)-3', RAD14, ... | | 著者 | Simon, N, Ebert, C, Schneider, S. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis for Bulky Adduct DNA Lesion Recognition by the Nucleotide Excision Repair Protein Rad14.
Chemistry, 22, 2016
|
|
7C0G
 
 | |
8OET
 
 | | SFX structure of the class II photolyase complexed with a thymine dimer | | 分子名称: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, DNA (14-mer), Deoxyribodipyrimidine photo-lyase, ... | | 著者 | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | 登録日 | 2023-03-12 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
3OOR
 
 | | I-SceI mutant (K86R/G100T)complexed with C/G+4 DNA substrate | | 分子名称: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | 著者 | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | 登録日 | 2010-08-31 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
5G32
 
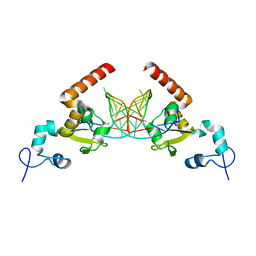 | | Structure of Rad14 in complex with acetylaminophenyl-guanine containing DNA | | 分子名称: | 5'-D(*GP*CP*TP*CP*TP*AP*6FKP*TP*CP*AP*TP*CP*AP*CP)-3', 5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*GP)-3', RAD14, ... | | 著者 | Simon, N, Ebert, C, Schneider, S. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for Bulky Adduct DNA Lesion Recognition by the Nucleotide Excision Repair Protein Rad14.
Chemistry, 22, 2016
|
|
4D9Y
 
 | | The crystal structure of Chelerythrine bound to DNA d(CGTACG) | | 分子名称: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, CALCIUM ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | 著者 | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | 登録日 | 2012-01-12 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of Chelerythrine bound to DNA d(CGTACG)
To be Published
|
|
4D9X
 
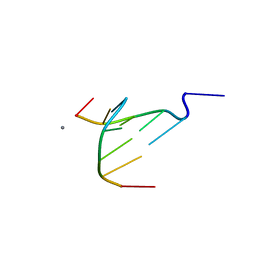 | | The crystal structure of Coptisine bound to DNA d(CGTACG) | | 分子名称: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, CALCIUM ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | 著者 | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | 登録日 | 2012-01-12 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | The crystal structure of Coptisine bound to DNA d(CGTACG)
to be published
|
|
3WI3
 
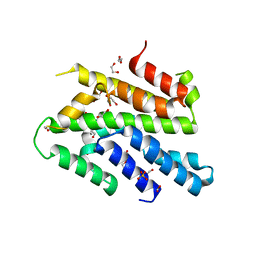 | | Crystal Structure of the Sld3/Treslin domain from yeast Sld3 | | 分子名称: | 1,2-ETHANEDIOL, DNA replication regulator SLD3, SULFATE ION | | 著者 | Itou, H, Araki, H, Shirakihara, Y. | | 登録日 | 2013-09-05 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the homology domain of the eukaryotic DNA replication proteins sld3/treslin.
Structure, 22, 2014
|
|
8V4T
 
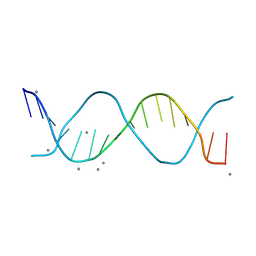 | |
6BQ9
 
 | | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida | | 分子名称: | CHLORIDE ION, DNA topoisomerase 4 subunit A, SODIUM ION | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-11-27 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida.
To Be Published
|
|
5N41
 
 | | Archaeal DNA polymerase holoenzyme - SSO6202 at 1.35 Ang resolution | | 分子名称: | 1,2-ETHANEDIOL, PolB1 Binding Protein 2 | | 著者 | Yan, J, Beattie, T.R, Rojas, A.L, Schermerhorn, K, Gristwood, T, Trinidad, J.C, Albers, S.V, Roversi, P, Gardner, A.F, Abrescia, N.G.A, Bell, S.D. | | 登録日 | 2017-02-09 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.351 Å) | | 主引用文献 | Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme.
Nat Commun, 8, 2017
|
|
