6OXU
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-99 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{({4-[(1R)-1-hydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.861 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6UBE
 
 | | Azide-triggered subtilisin SUBT_BACAM complexed with the peptide LFRAL | | 分子名称: | AZIDE ION, GLYCEROL, Peptide LFRAL, ... | | 著者 | Toth, E.A, Bryan, P.N, Orban, J, Gallagher, D.T, Custer, G. | | 登録日 | 2019-09-11 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6OY2
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-25 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-ethylbutyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXZ
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-20 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.961 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OYE
 
 | |
6UC8
 
 | | Guanine riboswitch bound to 8-aminoguanine | | 分子名称: | 8-AMINOGUANINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | 著者 | Matyjasik, M.M, Hall, S.D, Batey, R.T. | | 登録日 | 2019-09-15 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | High Affinity Binding of N2-Modified Guanine Derivatives Significantly Disrupts the Ligand Binding Pocket of the Guanine Riboswitch.
Molecules, 25, 2020
|
|
4ZU6
 
 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus influenzae in complex with pre-reactive o-acetyl serine, alpha-aminoacrylate reaction intermediate and Peptide inhibitor at the resolution of 2.25A | | 分子名称: | C-terminal peptide from Serine acetyltransferase, Cysteine synthase, O-ACETYLSERINE, ... | | 著者 | Ekka, M.K, Singh, A.K, Kaushik, A, Kumaran, S. | | 登録日 | 2015-05-15 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus inuen-zae in complex with pre-reactive o-acetyl serine and peptide inhibitor
To Be Published
|
|
4ZU1
 
 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus influenzae in complex with O-acetyl serine and peptide inhibitor | | 分子名称: | C-terminal peptide from Serine acetyltransferase, Cysteine synthase, GLYCEROL, ... | | 著者 | Ekka, M.K, Singh, A.K, Kaushik, A, Kumaran, S. | | 登録日 | 2015-05-15 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus inuen-zae in complex with O-acetyl serine and peptide inhibitor
To Be Published
|
|
6OFL
 
 | | Crystal structure of green fluorescent protein (GFP); S65T, Y66(3-ClY); ih circular permutant (50-51) | | 分子名称: | Green fluorescent protein (GFP); S65T, Y66(3-ClY); ih circular permutant (50-51) | | 著者 | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6OXW
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-100 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[(2-ethylbutyl)({4-[(1R)-1-hydroxyethyl]phenyl}sulfonyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.982 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6TYX
 
 | |
6OZH
 
 | |
6UC9
 
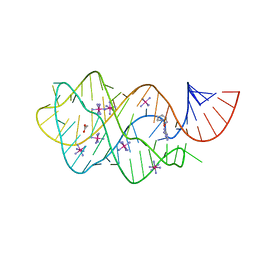 | |
6UCD
 
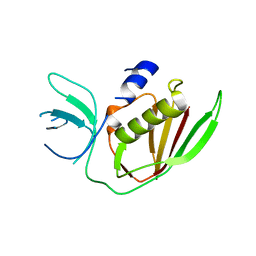 | |
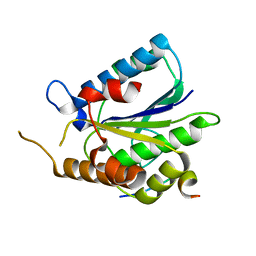
6TYU
 
 | |
6P2M
 
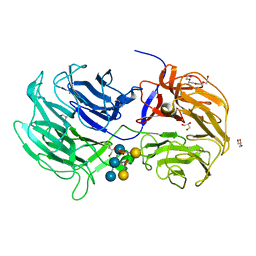 | | Crystal structure of the Caldicellulosiruptor lactoaceticus GH74 (ClGH74a) enzyme in complex with LLG xyloglucan | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Arnal, G. | | 登録日 | 2019-05-21 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
6U7D
 
 | |
6OZI
 
 | |
6U50
 
 | | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) | | 著者 | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | 登録日 | 2019-08-26 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
6U58
 
 | | Toho1 Beta Lactamase Glu166Gln Mutant | | 分子名称: | Beta-lactamase, SULFATE ION | | 著者 | Langan, P.S, Sullivan, B, Weiss, K.L. | | 登録日 | 2019-08-27 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | 主引用文献 | Probing the Role of the Conserved Residue Glu166 in a Class A Beta-Lactamase Using Neutron and X-ray Protein Crystallography
Acta Crystallogr.,Sect.D, 76, 2020
|
|
6PLX
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, desensitized state | | 分子名称: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yu, J, Zhu, H, Gouaux, E. | | 登録日 | 2019-07-01 | | 公開日 | 2021-01-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6P2H
 
 | |
6PGT
 
 | | Protein Tyrosine Phosphatase 1B (1-301), T177A mutant, vanadate bound state | | 分子名称: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, VANADATE ION | | 著者 | Cui, D.S, Lipchock, J.M, Loria, J.P. | | 登録日 | 2019-06-24 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
6PH1
 
 | |
6PHA
 
 | | Protein Tyrosine Phosphatase 1B (1-301), F182A mutant, vanadate bound state | | 分子名称: | ACETATE ION, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, ... | | 著者 | Cui, D.S, Lipchock, J.M, Loria, J.P. | | 登録日 | 2019-06-25 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
