7M1W
 
 | |
7LC8
 
 | | SARS-CoV-2 spike Protein TM domain | | 分子名称: | Spike protein S2' | | 著者 | Fu, Q, Chou, J.J. | | 登録日 | 2021-01-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Trimeric Hydrophobic Zipper Mediates the Intramembrane Assembly of SARS-CoV-2 Spike.
J.Am.Chem.Soc., 143, 2021
|
|
7LQT
 
 | | Solution NMR structure of the PNUTS amino-terminal Domain fused to Myc Homology Box 0 | | 分子名称: | Serine/threonine-protein phosphatase 1 regulatory subunit 10,Myc proto-oncogene protein fusion | | 著者 | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | 登録日 | 2021-02-15 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
7MDH
 
 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | 分子名称: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | 著者 | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | 登録日 | 1999-02-16 | | 公開日 | 1999-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
7M4R
 
 | |
7M5H
 
 | | Crystal structure of conserved protein from Enterococcus faecalis V583 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase | | 著者 | Nocek, B, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2021-03-23 | | 公開日 | 2021-04-07 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of conserved protein from Enterococcus faecalis V583
To Be Published
|
|
7MBK
 
 | |
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | 分子名称: | Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | 登録日 | 2021-04-06 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7M6B
 
 | | The Crystal Structure of Mcbe1 | | 分子名称: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | 著者 | Alahuhta, P.M, Lunin, V.V. | | 登録日 | 2021-03-25 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
7MC3
 
 | | Solution structure of Miz-1 zinc finger 12 | | 分子名称: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | 著者 | Boisvert, O, Lavigne, P. | | 登録日 | 2021-04-01 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7MC1
 
 | | Solution structure of Miz-1 Zinc finger 10 | | 分子名称: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | 著者 | Boisvert, O, Lavigne, P. | | 登録日 | 2021-04-01 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7MEU
 
 | |
4OFP
 
 | | Crystal Structure of SYG-2 D3-D4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-2 | | 著者 | Ozkan, E, Garcia, K.C. | | 登録日 | 2014-01-15 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
7MFP
 
 | | crystal structure of the L136 aminotransferase K185A from acanthamoeba polyphaga mimivirus in the presence of the UDP-viosamine external aldimine | | 分子名称: | (2R,3R,4S,5S,6R)-3,4-dihydroxy-5-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-methyloxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | 登録日 | 2021-04-10 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFQ
 
 | | crystal structure of the L136 aminotransferase from acanthamoeba polyphaga mimivirus in complex with the TDP-viosamine external aldimine | | 分子名称: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | 登録日 | 2021-04-10 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MC2
 
 | |
7MJR
 
 | | Vip4Da2 toxin structure | | 分子名称: | CALCIUM ION, SULFATE ION, Vip4Da1 protein | | 著者 | Rydel, T.J, Duda, D, Zheng, M, Henry, A. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.22 Å) | | 主引用文献 | Structural and functional insights into the first Bacillus thuringiensis vegetative insecticidal protein of the Vpb4 fold, active against western corn rootworm.
Plos One, 16, 2021
|
|
7ML9
 
 | | The Mpp75Aa1.1 beta-pore-forming protein from Brevibacillus laterosporus | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Insecticidal protein, ... | | 著者 | Rydel, T.J, Zheng, M, Evdokimov, A. | | 登録日 | 2021-04-27 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural and functional characterization of Mpp75Aa1.1, a putative beta-pore forming protein from Brevibacillus laterosporus active against the western corn rootworm.
Plos One, 16, 2021
|
|
7MHF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
5KO6
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with cytosine and ribose-1-phosphate | | 分子名称: | 1-O-phosphono-alpha-D-ribofuranose, 6-AMINOPYRIMIDIN-2(1H)-ONE, Purine nucleoside phosphorylase | | 著者 | Torini, J.R, Romanello, L, Bird, L, Owens, R, Brandao-Neto, J, Pereira, H.M. | | 登録日 | 2016-06-29 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
7MHG
 
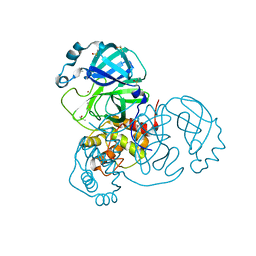 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 240 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5302 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7M3Q
 
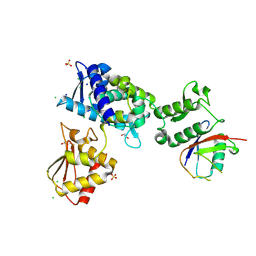 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | 分子名称: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | 著者 | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | 登録日 | 2021-03-18 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
7MHK
 
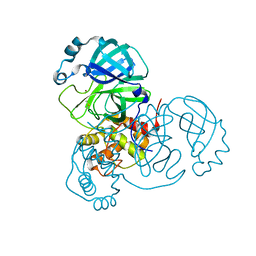 | | Crystal Structure of Apo/Unliganded SARS-CoV-2 Main Protease (Mpro) at 310 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9601 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHJ
 
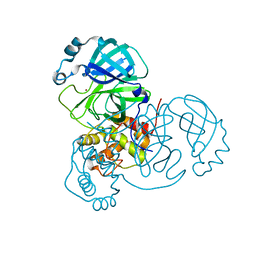 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K and High Humidity | | 分子名称: | 3C-like proteinase, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.0005 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MJN
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
