3A7D
 
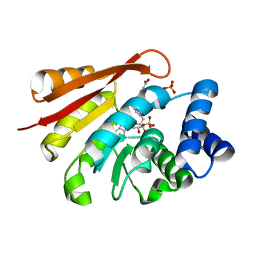 | | Crystal Structures of rat Catechol-O-Methyltransferase complexed with new bi-substrate type inhibitor | | 分子名称: | 5'-deoxy-5'-[4-({[(2,3-dihydroxy-5-nitrophenyl)carbonyl]amino}methyl)-1H-1,2,3-triazol-1-yl]adenosine, Catechol O-methyltransferase, MAGNESIUM ION, ... | | 著者 | Tsuji, E. | | 登録日 | 2009-09-26 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Hit to Lead: Comprehensive Strategy of de novo Scaffold Generation by FBDD. Part 2: Ligand Fishing using Mass Spectrometry by Detection of Ligand-Protein Non-Covalent Complex after Matrix Click Chemistry
To be Published
|
|
3A7E
 
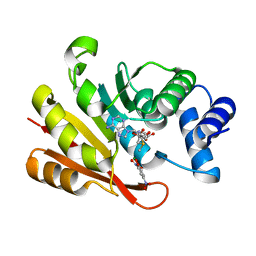 | | Crystal structure of human COMT complexed with SAM and 3,5-dinitrocatechol | | 分子名称: | 3,5-DINITROCATECHOL, Catechol O-methyltransferase, MAGNESIUM ION, ... | | 著者 | Tsuji, E. | | 登録日 | 2009-09-26 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Hit to Lead: Comprehensive Strategy of de novo Scaffold Generation by FBDD. Part 1: In silico Fragments Linking and Verification of Spatial Proximity using Inter Ligand NOE Approachs
To be Published
|
|
1I4U
 
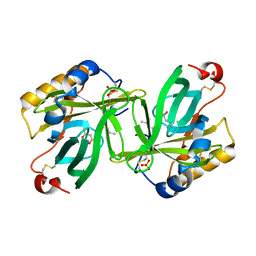 | | THE C1 SUBUNIT OF ALPHA-CRUSTACYANIN | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CRUSTACYANIN, SULFATE ION | | 著者 | Gordon, E.J, Leonard, G.A, McSweeney, S, Zagalsky, P.F. | | 登録日 | 2001-02-23 | | 公開日 | 2001-09-19 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | The C1 subunit of alpha-crustacyanin: the de novo phasing of the crystal structure of a 40 kDa homodimeric protein using the anomalous scattering from S atoms combined with direct methods.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5LY2
 
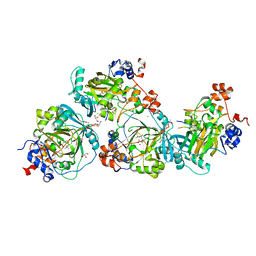 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Macrocyclic PEPTIDE Inhibitor CP2_R6Kme3 (13-mer) | | 分子名称: | CHLORIDE ION, CP2_R6Kme3, GLYCEROL, ... | | 著者 | Chowdhury, R, Madden, S.K, Hopkinson, R, Schofield, C.J. | | 登録日 | 2016-09-23 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | 著者 | Doyle, L.A, Stoddard, B.L. | | 登録日 | 2019-04-05 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
8VSW
 
 | | 4-MERCAPTOPHENOL-ALPHA3C | | 分子名称: | 4-mercaptophenol-alpha3C protein, 4-sulfanylphenol | | 著者 | Tommos, C. | | 登録日 | 2024-01-24 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Switching the proton-coupled electron transfer mechanism for non-canonical tyrosine residues in a de novo protein.
Chem Sci, 15, 2024
|
|
5UOI
 
 | | Solution structure of the de novo mini protein HHH_rd1_0142 | | 分子名称: | HHH_rd1_0142 | | 著者 | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | 登録日 | 2017-01-31 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | 分子名称: | HEEH_rd4_0097 | | 著者 | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | 登録日 | 2017-02-24 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
4F2V
 
 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | 分子名称: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | 著者 | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-05-08 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.493 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
8E1E
 
 | |
5UP1
 
 | | Solution structure of the de novo mini protein EEHEE_rd3_1049 | | 分子名称: | EEHEE_rd3_1049 | | 著者 | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | 登録日 | 2017-02-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP5
 
 | | Solution structure of the de novo mini protein EHEE_rd1_0284 | | 分子名称: | EHEE_rd1_0284 | | 著者 | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | 登録日 | 2017-02-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UZL
 
 | |
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
7UBH
 
 | |
3Q7W
 
 | |
3Q7X
 
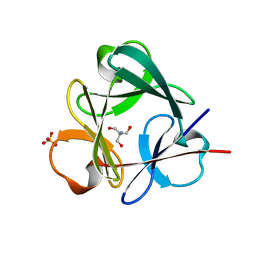 | | Crystal structure of Symfoil-4P/PV1: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 1 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo designed beta-trefoil architecture with symmetric primary structure | | 著者 | Blaber, M, Lee, J. | | 登録日 | 2011-01-05 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
