5WVK
 
 | | Yeast proteasome-ADP-AlFx | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Ding, Z, Cong, Y. | | 登録日 | 2016-12-25 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
7ZI4
 
 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | 登録日 | 2022-04-07 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | 分子名称: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | 著者 | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | 登録日 | 2018-02-10 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
7KSO
 
 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | 分子名称: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | 著者 | Grau, D.J, Armache, K.J. | | 登録日 | 2020-11-23 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7AT8
 
 | | Histone H3 recognition by nucleosome-bound PRC2 subunit EZH2. | | 分子名称: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | 著者 | Finogenova, K, Benda, C, Schaefer, I.B, Poepsel, S, Strauss, M, Mueller, J. | | 登録日 | 2020-10-29 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Elife, 9, 2020
|
|
4Y72
 
 | | Human CDK1/CyclinB1/CKS2 With Inhibitor | | 分子名称: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | 著者 | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | 登録日 | 2015-02-13 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
4YC3
 
 | | CDK1/CyclinB1/CKS2 Apo | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | 著者 | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | 登録日 | 2015-02-19 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
7PFX
 
 | |
7PEX
 
 | |
7PFC
 
 | |
7PEZ
 
 | |
7PF3
 
 | |
7PF5
 
 | |
7PET
 
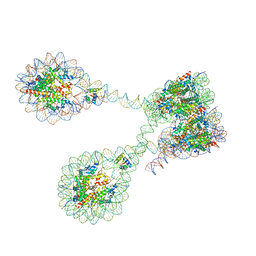 | | The 4x177 nucleosome array containing H1 | | 分子名称: | DNA (702-MER), Histone H1.4, Histone H2A type 1-B/E, ... | | 著者 | Dombrowski, M, Cramer, P. | | 登録日 | 2021-08-11 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Histone H1 binding to nucleosome arrays depends on linker DNA length and trajectory.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PF6
 
 | |
7PFU
 
 | |
7PF2
 
 | |
7PFA
 
 | |
7PEU
 
 | |
7PFD
 
 | |
7PFE
 
 | |
7PFT
 
 | |
7PF0
 
 | |
7PFV
 
 | |
7PFW
 
 | |
