8X8S
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | 分子名称: | Polyhedrin,Myc proto-oncogene protein | | 著者 | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | 登録日 | 2023-11-28 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7Z
 
 | |
8X7X
 
 | |
8X7K
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination) | | 分子名称: | DNA (143-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | 登録日 | 2023-11-24 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7J
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy | | 分子名称: | DNA (144-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | 登録日 | 2023-11-24 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7I
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | 登録日 | 2023-11-24 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X79
 
 | | MRE-269 bound Prostacyclin Receptor G protein complex | | 分子名称: | 2-[4-[(5,6-diphenylpyrazin-2-yl)-propan-2-yl-amino]butoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | 登録日 | 2023-11-23 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
8X77
 
 | | Enterovirus proteinase with host factor | | 分子名称: | 2A protein, Actin-histidine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Gao, X, Cui, S. | | 登録日 | 2023-11-23 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.52 Å) | | 主引用文献 | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
8X73
 
 | | Crystal structure of Peroxiredoxin I in complex with compound 19-069 | | 分子名称: | Peroxiredoxin-1, methyl (2~{S})-2-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]-3-oxidanyl-propanoate | | 著者 | Zhang, H, Luo, C. | | 登録日 | 2023-11-22 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8X6Z
 
 | |
8X6R
 
 | | KRasG12C in complex with inhibitor | | 分子名称: | 1-[7-[6-ethenyl-8-ethoxy-7-(5-methyl-1~{H}-indazol-4-yl)-2-(1-methylpiperidin-4-yl)oxy-quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Amano, Y, Tateishi, Y. | | 登録日 | 2023-11-21 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of ASP6918, a KRAS G12C inhibitor: Synthesis and structure-activity relationships of 1-{2,7-diazaspiro[3.5]non-2-yl}prop-2-en-1-one derivatives as covalent inhibitors with good potency and oral activity for the treatment of solid tumors.
Bioorg.Med.Chem., 98, 2023
|
|
8X6M
 
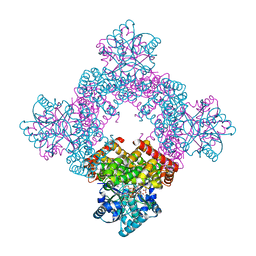 | | Crystal Structure of Glycerol Dehydrogenase in the Presence of NAD+ and Glycerol | | 分子名称: | GLYCEROL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Park, T, Kang, J.Y, Jin, M, Yang, J, Kim, H, Noh, C, Eom, S.H. | | 登録日 | 2023-11-21 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into the octamerization of glycerol dehydrogenase.
Plos One, 19, 2024
|
|
8X61
 
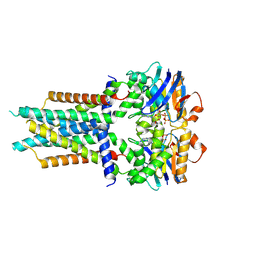 | | Cryo-EM structure of ATP-bound FtsE(E163Q)X | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX | | 著者 | Zhang, Z.Y, Chen, Y.T. | | 登録日 | 2023-11-20 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
8X5V
 
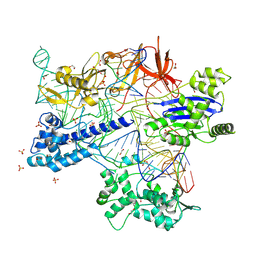 | | BlCas9-sgRNA-target DNA complex | | 分子名称: | 1,2-ETHANEDIOL, BlCas9, CHLORIDE ION, ... | | 著者 | Nakane, T, Nakagawa, R, Yamashita, K, Nishimasu, H, Nureki, O. | | 登録日 | 2023-11-19 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and engineering of Brevibacillus laterosporus Cas9.
Commun Biol, 7, 2024
|
|
8X5R
 
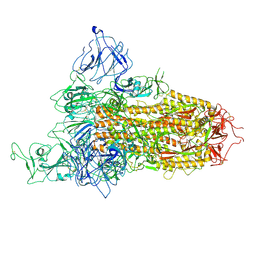 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-17 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5Q
 
 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-17 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5M
 
 | | The Crystal Structure of JNK1 from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, 3-[4-(dimethylamino)butanoylamino]-~{N}-[3-methyl-4-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl]benzamide, GLYCEROL, ... | | 著者 | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | 登録日 | 2023-11-17 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Crystal Structure of JNK1 from Biortus.
To Be Published
|
|
8X5L
 
 | | The Crystal Structure of PRKACA from Biortus. | | 分子名称: | (2S)-2-(4-chlorophenyl)-2-hydroxy-2-[4-(1H-pyrazol-4-yl)phenyl]ethanaminium, SODIUM ION, cAMP-dependent protein kinase catalytic subunit alpha | | 著者 | Wang, F, Cheng, W, Lv, Z, Lin, D, Pan, W. | | 登録日 | 2023-11-17 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Crystal Structure of PRKACA from Biortus.
To Be Published
|
|
8X5E
 
 | | Cryo-EM structure of human XPR1 in open state | | 分子名称: | PHOSPHATE ION, Solute carrier family 53 member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Jiang, D.H, Yan, R. | | 登録日 | 2023-11-17 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | Human XPR1 structures reveal phosphate export mechanism.
Nature, 2024
|
|
8X56
 
 | | BA.2.86 Spike Trimer with T356K mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X55
 
 | | BA.2.86 Spike Trimer with T356K mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X50
 
 | | BA.2.86 Spike Trimer with ins483V mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X4Z
 
 | | BA.2.86 Spike Trimer with ins483V mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X4L
 
 | |
8X4H
 
 | | SARS-CoV-2 JN.1 Spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-15 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
