6E3L
 
 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, ... | | 著者 | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | 登録日 | 2018-07-14 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
5IEY
 
 | | Crystal structure of a CDK inhibitor bound to CDK2 | | 分子名称: | 4-[(4-{[(2R,3R)-3-hydroxybutan-2-yl]amino}pyrimidin-2-yl)amino]benzene-1-sulfonamide, Cyclin-dependent kinase 2 | | 著者 | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | 登録日 | 2016-02-25 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5GW9
 
 | |
5GPD
 
 | |
5X3F
 
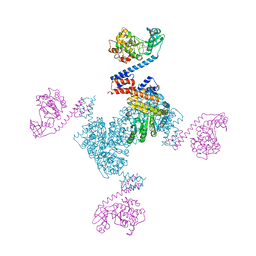 | | Crystal structure of the YgjG-Protein A-Zpa963-PKA catalytic domain | | 分子名称: | Putrescine aminotransferase,Immunoglobulin G-binding protein A, Zpa963,cAMP-dependent protein kinase catalytic subunit alpha | | 著者 | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | 登録日 | 2017-02-05 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.38 Å) | | 主引用文献 | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5GRS
 
 | | Complex structure of the fission yeast SREBP-SCAP binding domains | | 分子名称: | Sterol regulatory element-binding protein 1, Sterol regulatory element-binding protein cleavage-activating protein | | 著者 | Gong, X, Qian, H.W, Wu, J.P, Yan, N. | | 登録日 | 2016-08-12 | | 公開日 | 2016-12-14 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (5.4 Å) | | 主引用文献 | Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization
Cell Res., 26, 2016
|
|
5ILU
 
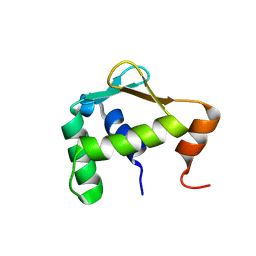 | | Autoinhibited ETV4 | | 分子名称: | ETS translocation variant 4 | | 著者 | Whitby, F.G, Currie, S.L. | | 登録日 | 2016-03-04 | | 公開日 | 2017-02-22 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
6EYC
 
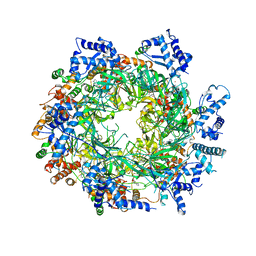 | | Re-refinement of the MCM2-7 double hexamer using ISOLDE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | 著者 | Croll, T.I. | | 登録日 | 2017-11-11 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3L2O
 
 | |
1XFV
 
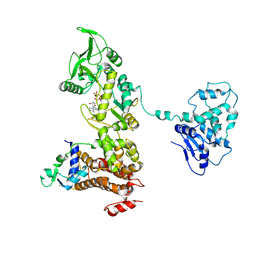 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | 分子名称: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | 著者 | Shen, Q, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | 登録日 | 2004-09-15 | | 公開日 | 2005-05-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFW
 
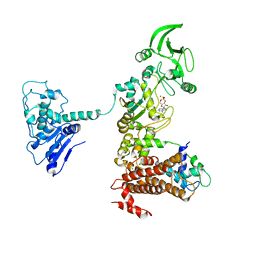 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3'5' cyclic AMP (cAMP) | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | 著者 | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | 登録日 | 2004-09-15 | | 公開日 | 2005-05-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
2ZOQ
 
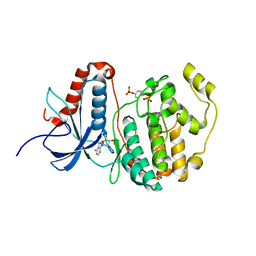 | | Structural dissection of human mitogen-activated kinase ERK1 | | 分子名称: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 3, SODIUM ION, ... | | 著者 | Kinoshita, T, Tada, T, Nakae, S, Yoshida, I. | | 登録日 | 2008-06-01 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Crystal structure of human mono-phosphorylated ERK1 at Tyr204
Biochem.Biophys.Res.Commun., 377, 2008
|
|
6S9S
 
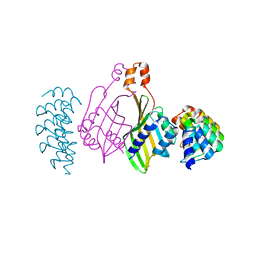 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 10 | | 分子名称: | Darpin 10, LIM domain-binding protein 1 | | 著者 | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | 登録日 | 2019-07-15 | | 公開日 | 2019-10-09 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3X2V
 
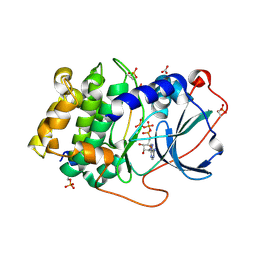 | | Michaelis-like complex of cAMP-dependent Protein Kinase Catalytic Subunit | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | 著者 | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | 登録日 | 2015-01-02 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
6SJA
 
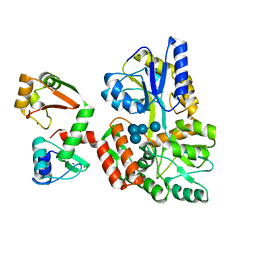 | | Structure of HPV16 E6 oncoprotein in complex with IRF3 LxxLL motif | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | 著者 | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | 登録日 | 2019-08-13 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Deciphering de molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
6SVL
 
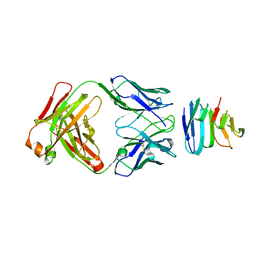 | |
6SXB
 
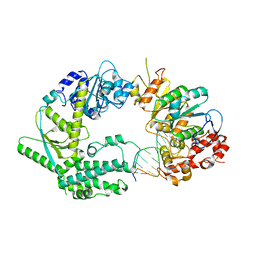 | | XPF-ERCC1 Cryo-EM Structure, DNA-Bound form | | 分子名称: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*AP*TP*GP*CP*TP*GP*A)-3'), DNA excision repair protein ERCC-1, ... | | 著者 | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | 登録日 | 2019-09-25 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (7.9 Å) | | 主引用文献 | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
7V7C
 
 | | CryoEM structure of DDB1-VprBP-Vpr-UNG2(94-313) complex | | 分子名称: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, Protein Vpr, ... | | 著者 | Wang, D, Xu, J, Liu, Q, Xiang, Y. | | 登録日 | 2021-08-21 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into the HIV-1 Vpr mediated ubiquitination through the Cullin-RING E3 ubiquitin ligase
To Be Published
|
|
3X2U
 
 | | Michaelis-like initial complex of cAMP-dependent Protein Kinase Catalytic Subunit. | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Substrate Peptide, ... | | 著者 | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | 登録日 | 2015-01-02 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
6SVK
 
 | | human Myeloid-derived growth factor (MYDGF) | | 分子名称: | MALONATE ION, Myeloid-derived growth factor | | 著者 | Ebenhoch, R, Nar, H. | | 登録日 | 2019-09-18 | | 公開日 | 2019-11-27 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.604 Å) | | 主引用文献 | Crystal structure and receptor-interacting residues of MYDGF - a protein mediating ischemic tissue repair.
Nat Commun, 10, 2019
|
|
7V7B
 
 | |
6SLX
 
 | | Fragment AZ-010 binding at the TAZpS89/14-3-3 sigma interface | | 分子名称: | 14-3-3 protein sigma, TAZpS89, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]propanamide | | 著者 | Ottmann, C, Wolter, M, Guillory, X, Genet, S, Somsen, B, Leysen, S, Castaldi, P. | | 登録日 | 2019-08-20 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.800045 Å) | | 主引用文献 | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
7VD4
 
 | | Crystal structure of BPTF-BRD with ligand TP248 bound | | 分子名称: | 6-[4-[3-(dimethylamino)propoxy]phenyl]-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | 著者 | Lu, T, Lu, H.B. | | 登録日 | 2021-09-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85659146 Å) | | 主引用文献 | Discovery of a highly potent CECR2 bromodomain inhibitor with 7H-pyrrolo[2,3-d] pyrimidine scaffold.
Bioorg.Chem., 123, 2022
|
|
6SQ4
 
 | |
6SQ3
 
 | |
