6PF0
 
 | |
6PEZ
 
 | |
6PX7
 
 | |
6PX8
 
 | |
7VS4
 
 | | Crystal structure of PacII_M1M2S-DNA(m6A)-SAH complex | | 分子名称: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | 著者 | Zhu, J, Gao, P. | | 登録日 | 2021-10-25 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
7VRU
 
 | | Crystal structure of PacII_M1M2S-DNA-SAH complex | | 分子名称: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | 著者 | Zhu, J, Gao, P. | | 登録日 | 2021-10-25 | | 公開日 | 2022-11-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
1B4G
 
 | | CONTROL OF K+ CHANNEL GATING BY PROTEIN PHOSPHORYLATION: STRUCTURAL SWITCHES OF THE INACTIVATION GATE, NMR, 22 STRUCTURES | | 分子名称: | POTASSIUM CHANNEL | | 著者 | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | 登録日 | 1998-12-22 | | 公開日 | 1999-04-27 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
1B4I
 
 | | Control of K+ Channel Gating by protein phosphorylation: structural switches of the inactivation gate, NMR, 22 structures | | 分子名称: | POTASSIUM CHANNEL | | 著者 | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | 登録日 | 1998-12-22 | | 公開日 | 1999-04-27 | | 最終更新日 | 2022-03-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
1E3M
 
 | | The crystal structure of E. coli MutS binding to DNA with a G:T mismatch | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP* GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP* TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Lamers, M.H, Perrakis, A, Enzlin, J.H, Winterwerp, H.H.K, De Wind, N, Sixma, T.K. | | 登録日 | 2000-06-19 | | 公開日 | 2000-11-01 | | 最終更新日 | 2017-07-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Nature, 407, 2000
|
|
1D2N
 
 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | 分子名称: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | 著者 | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | 登録日 | 1998-06-30 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
6D0Q
 
 | |
6D0R
 
 | |
4CS5
 
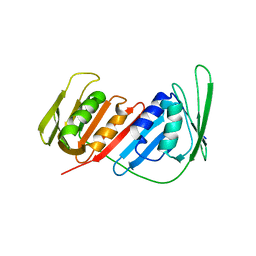 | | Crystal Structure of PCNA from Litopenaeus vannamei | | 分子名称: | PROLIFERATING CELL NUCLEAR ANTIGEN | | 著者 | Carrasco-Miranda, J.S, Lopez-Zavala, A.A, De-La-Mora, E, Rudino-Pinera, E, Brieba, L.G, Sotelo-Mundo, R.R. | | 登録日 | 2014-03-04 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of the Shrimp Proliferating Cell Nuclear Antigen: Structural Complementarity with Wssv DNA Polymerase Pip-Box.
Plos One, 9, 2014
|
|
6DEI
 
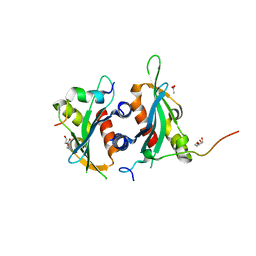 | | Structure of Dse3-Csm1 complex | | 分子名称: | ACETATE ION, Monopolin complex subunit CSM1, Protein DSE3, ... | | 著者 | Singh, N, Corbett, K.D. | | 登録日 | 2018-05-12 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | The budding-yeast RWD protein Csm1 scaffolds diverse protein complexes through a conserved structural mechanism.
Protein Sci., 27, 2018
|
|
5V03
 
 | |
5T0Q
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | 分子名称: | Protein TIFY 9, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | 登録日 | 2016-08-16 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T0F
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | 分子名称: | Protein TIFY 9, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | 登録日 | 2016-08-16 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V02
 
 | |
7S7P
 
 | |
7KZL
 
 | |
4DAM
 
 | | Crystal structure of small single-stranded DNA-binding protein from Streptomyces coelicolor | | 分子名称: | Single-stranded DNA-binding protein 1 | | 著者 | Filic, Z, Herron, P, Ivic, N, Luic, M, Manjasetty, B.A, Paradzik, T, Vujaklija, D. | | 登録日 | 2012-01-13 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-function relationships of two paralogous single-stranded DNA-binding proteins from Streptomyces coelicolor: implication of SsbB in chromosome segregation during sporulation.
Nucleic Acids Res., 41, 2013
|
|
4GZZ
 
 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | 登録日 | 2012-09-06 | | 公開日 | 2013-02-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (4.2927 Å) | | 主引用文献 | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
4GZY
 
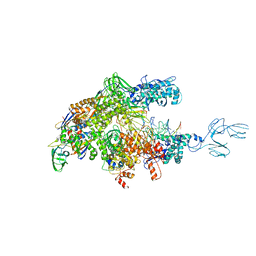 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | 登録日 | 2012-09-06 | | 公開日 | 2013-02-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.5054 Å) | | 主引用文献 | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
5WCV
 
 | |
7DW5
 
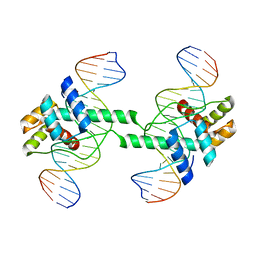 | | Crystal structure of DUX4 HD1-HD2 domain complexed with ERG sites | | 分子名称: | BROMIDE ION, DNA (5'-D(P*CP*GP*AP*CP*TP*TP*GP*AP*TP*GP*AP*GP*AP*TP*TP*AP*GP*AP*CP*TP*G)-3'), Double homeobox protein 4-like protein 2 | | 著者 | Zhang, H, Cheng, N, Li, Z, Zhang, W, Dong, X, Huang, J, Meng, G. | | 登録日 | 2021-01-15 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia.
Cancer Commun (Lond), 41, 2021
|
|
