3NMO
 
 | |
4CLA
 
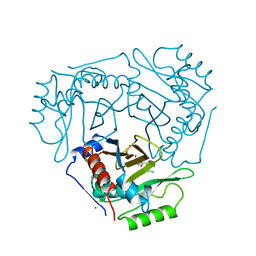 | |
7JJU
 
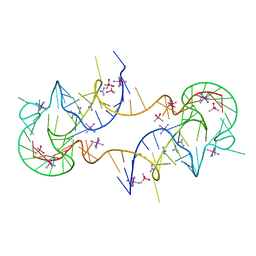 | | Crystal structure of en exoribonuclease-resistant RNA (xrRNA) from Potato leafroll virus (PLRV) | | 分子名称: | CACODYLATE ION, Guanidinium, IRIDIUM HEXAMMINE ION, ... | | 著者 | Steckelberg, A.-L, Vicens, Q, Auffinger, P, Costantino, D.C, Nix, J.C, Kieft, J.S. | | 登録日 | 2020-07-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.604 Å) | | 主引用文献 | The crystal structure of a Polerovirus exoribonuclease-resistant RNA shows how diverse sequences are integrated into a conserved fold.
Rna, 26, 2020
|
|
4BZ4
 
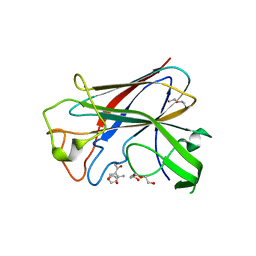 | | CorA is a surface-associated copper-binding protein important in Methylomicrobium album BG8 copper acquisition | | 分子名称: | CALCIUM ION, COPPER (I) ION, COPPER-REPRESSIBLE POLYPEPTIDE, ... | | 著者 | Johnson, K.A, Ve, T, Pedersen, R.B, Lillehaug, J.R, Jensen, H.B, Helland, R, Karlsen, O.A. | | 登録日 | 2013-07-24 | | 公開日 | 2014-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Cora is a Copper Repressible Surface-Associated Copper(I)-Binding Protein Produced in Methylomicrobium Album Bg8.
Plos One, 9, 2014
|
|
3PDY
 
 | |
7K18
 
 | | Cardiac Sodium channel with toxin bound | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-like toxin Lqh3, ... | | 著者 | Jiang, D, Catterall, W.A. | | 登録日 | 2020-09-07 | | 公開日 | 2021-01-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for voltage-sensor trapping of the cardiac sodium channel by a deathstalker scorpion toxin.
Nat Commun, 12, 2021
|
|
7XYH
 
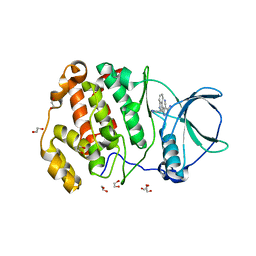 | | Crystal structure of CK2a2 complexed with AG1112 | | 分子名称: | 1,2-ETHANEDIOL, 5-azanyl-3-[(~{Z})-1-cyano-2-(1~{H}-indol-3-yl)ethenyl]-1~{H}-pyrazole-4-carbonitrile, Casein kinase II subunit alpha' | | 著者 | Ikeda, A, Kinoshita, T, Tsuyuguchi, M. | | 登録日 | 2022-06-01 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Bivalent binding mode of an amino-pyrazole inhibitor indicates the potentials for CK2 alpha 1-selective inhibitors.
Biochem.Biophys.Res.Commun., 630, 2022
|
|
6DG2
 
 | |
6E8M
 
 | |
7X73
 
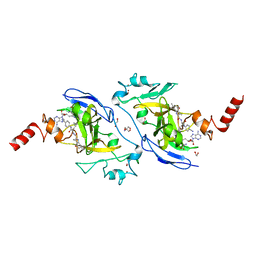 | | Structure of G9a in complex with RK-701 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | 著者 | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Shirouzu, M, Umehara, T. | | 登録日 | 2022-03-09 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nat Commun, 14, 2023
|
|
5M7O
 
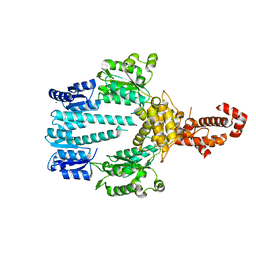 | | Crystal structure of NtrX from Brucella abortus processed with the CrystalDirect automated mounting and cryo-cooling technology | | 分子名称: | MAGNESIUM ION, Nitrogen assimilation regulatory protein | | 著者 | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | 登録日 | 2016-10-28 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
8WFX
 
 | | Cryo-EM structure of CRISPR-Csm effector complex from Mycobacterium canettii | | 分子名称: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | 著者 | Huo, Y, Ma, X, Jiang, T. | | 登録日 | 2023-09-20 | | 公開日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | Type-III-A structure of mycobacteria CRISPR-Csm complexes involving atypical crRNAs.
Int.J.Biol.Macromol., 260, 2024
|
|
3PY3
 
 | |
6ENL
 
 | |
4FG6
 
 | | Structure of EcCLC E148A mutant in Glutamate | | 分子名称: | Fab fragment (Heavy chain), Fab fragment (Light chain), H(+)/Cl(-) exchange transporter ClcA | | 著者 | Feng, L, MacKinnon, R. | | 登録日 | 2012-06-04 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.019 Å) | | 主引用文献 | Molecular mechanism of proton transport in CLC Cl-/H+ exchange transporters.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | 著者 | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | 登録日 | 2012-05-22 | | 公開日 | 2012-06-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.357 Å) | | 主引用文献 | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
4FIV
 
 | | FIV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | 分子名称: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, FELINE IMMUNODEFICIENCY VIRUS PROTEASE | | 著者 | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | 登録日 | 1998-07-15 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
8YBE
 
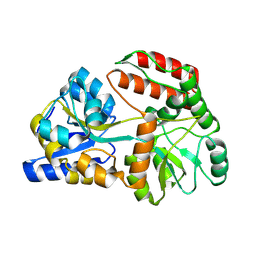 | |
1NH5
 
 | | AUTOMATIC ASSIGNMENT OF NMR DATA AND DETERMINATION OF THE PROTEIN STRUCTURE OF A NEW WORLD SCORPION NEUROTOXIN USING NOAH/DIAMOD | | 分子名称: | Neurotoxin 5 | | 著者 | Xu, Y, Jablonsky, M.J, Jackson, P.L, Krishna, N.R, Braun, W. | | 登録日 | 2002-12-18 | | 公開日 | 2003-01-07 | | 最終更新日 | 2018-01-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Automatic assignment of NOESY Cross peaks and determination of the protein structure of a new world scorpion neurotoxin Using NOAH/DIAMOD
J.Magn.Reson., 148, 2001
|
|
8XAX
 
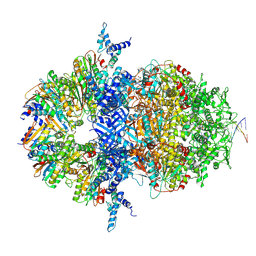 | |
8XAU
 
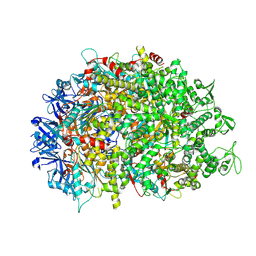 | | Cryo-EM structure of HerA | | 分子名称: | ATP-binding protein | | 著者 | Wang, Y, Deng, Z. | | 登録日 | 2023-12-05 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8XAV
 
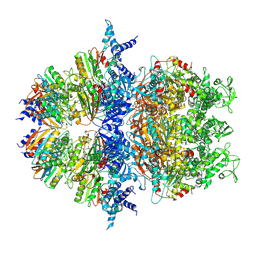 | |
8XAW
 
 | |
8Y0F
 
 | |
8XUR
 
 | | BA.2.86 Spike Trimer in complex with heparan sulfate | | 分子名称: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yue, C, Liu, P. | | 登録日 | 2024-01-14 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
