7O72
 
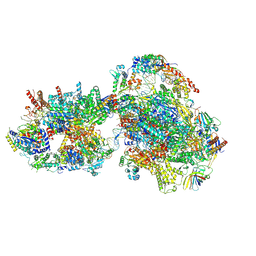 | | Yeast RNA polymerase II transcription pre-initiation complex with closed promoter DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-12 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O4K
 
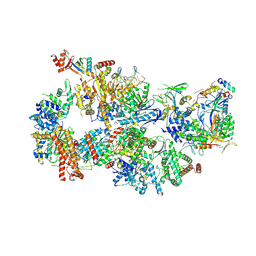 | | Yeast TFIIH in the contracted state within the pre-initiation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-06 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
9FFF
 
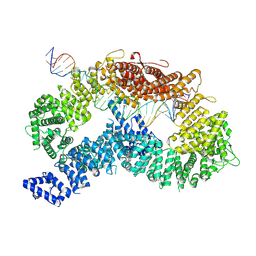 | | dsDNA-FANCD2-FANCI complex | | 分子名称: | DNA (32-MER), DNA (33-MER), Fanconi anemia complementation group I, ... | | 著者 | Alcon, P, Passmore, L.A. | | 登録日 | 2024-05-23 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions.
Nature, 632, 2024
|
|
9FFB
 
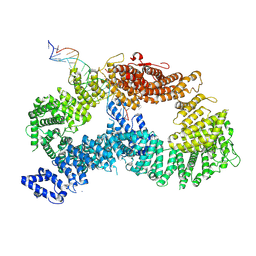 | | ss-dsDNA-FANCD2-FANCI complex | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*GP*TP*CP*TP*CP*TP*AP*GP*AP*CP*AP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*GP*CP*TP*GP*TP*CP*TP*AP*GP*AP*GP*AP*CP*AP*TP*CP*GP*AP*T)-3'), Fanconi anemia complementation group I, ... | | 著者 | Alcon, P, Passmore, L.A. | | 登録日 | 2024-05-22 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions.
Nature, 632, 2024
|
|
2NB8
 
 | |
2NB7
 
 | |
7OW8
 
 | | CryoEM structure of the ABC transporter BmrA E504A mutant in complex with ATP-Mg | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | 著者 | Gobet, A, Schoehn, G, Falson, P, Chaptal, V. | | 登録日 | 2021-06-17 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
3P87
 
 | | Structure of human PCNA bound to RNASEH2B PIP box peptide | | 分子名称: | Proliferating cell nuclear antigen, Ribonuclease H2 subunit B | | 著者 | Bubeck, D, Reijns, M.A, Graham, S.C, Astell, K.R, Jones, E.Y, Jackson, A.P. | | 登録日 | 2010-10-13 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | PCNA directs type 2 RNase H activity on DNA replication and repair substrates.
Nucleic Acids Res., 39, 2011
|
|
3PVS
 
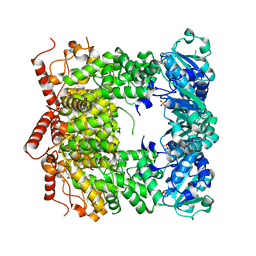 | | Structure and biochemical activities of Escherichia coli MgsA | | 分子名称: | PHOSPHATE ION, Replication-associated recombination protein A | | 著者 | Page, A.N, George, N.P, Marceau, A.H, Cox, M.M, Keck, J.L. | | 登録日 | 2010-12-07 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and Biochemical Activities of Escherichia coli MgsA.
J.Biol.Chem., 286, 2011
|
|
5DCA
 
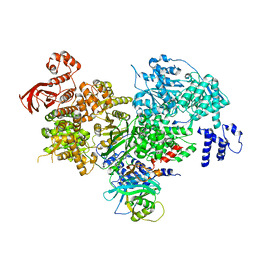 | | Crystal structure of yeast full length Brr2 in complex with Prp8 Jab1 domain | | 分子名称: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2 | | 著者 | Absmeier, E, Wollenhaupt, J, Santos, K.F, Wahl, M.C. | | 登録日 | 2015-08-23 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation.
Genes Dev., 29, 2015
|
|
3QQB
 
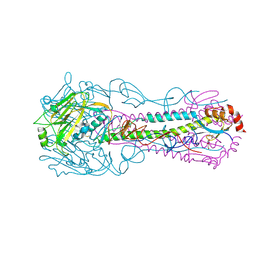 | | Crystal structure of HA2 R106H mutant of H2 hemagglutinin, neutral pH form | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xu, R, Wilson, I.A. | | 登録日 | 2011-02-15 | | 公開日 | 2011-03-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Characterization of an Early Fusion Intermediate of Influenza Virus Hemagglutinin.
J.Virol., 85, 2011
|
|
3QQE
 
 | | Crystal structure of HA2 R106H mutant of H2 hemagglutinin, re-neutralized form | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xu, R, Wilson, I.A. | | 登録日 | 2011-02-15 | | 公開日 | 2011-03-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Characterization of an Early Fusion Intermediate of Influenza Virus Hemagglutinin.
J.Virol., 85, 2011
|
|
3QQI
 
 | |
3QQO
 
 | |
4E0X
 
 | |
4EXP
 
 | | Structure of mouse Interleukin-34 in complex with mouse FMS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-34, Macrophage colony-stimulating factor 1 receptor | | 著者 | Liu, H, Leo, C, Chen, X, Wong, B.R, Williams, L.T, Lin, H, He, X. | | 登録日 | 2012-04-30 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1.
Biochim.Biophys.Acta, 1824, 2012
|
|
4WVE
 
 | |
7TDZ
 
 | |
6ITC
 
 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | 登録日 | 2018-11-21 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
6LDI
 
 | |
1LWJ
 
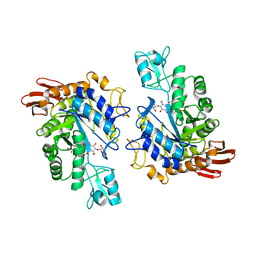 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE/ACARBOSE COMPLEX | | 分子名称: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, MODIFIED ACARBOSE PENTASACCHARIDE | | 著者 | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | 登録日 | 2002-05-31 | | 公開日 | 2002-08-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
1LWH
 
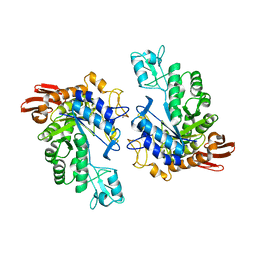 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE | | 分子名称: | 4-alpha-glucanotransferase, CALCIUM ION | | 著者 | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | 登録日 | 2002-05-31 | | 公開日 | 2002-07-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
3T0P
 
 | |
4M81
 
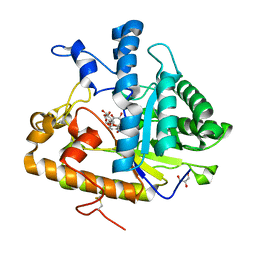 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution | | 分子名称: | 4-nitrophenyl beta-D-glucopyranoside, EXO-1,3-BETA-GLUCANASE, GLYCEROL, ... | | 著者 | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | 登録日 | 2013-08-12 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
4M82
 
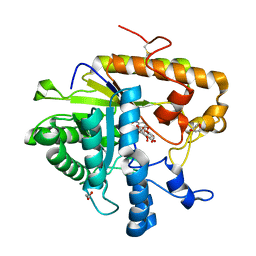 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with p-nitrophenyl-gentiobioside (product) at 1.6A resolution | | 分子名称: | 1,2-ETHANEDIOL, 4-nitrophenyl 6-O-beta-D-glucopyranosyl-beta-D-glucopyranoside, EXO-1,3-BETA-GLUCANASE, ... | | 著者 | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | 登録日 | 2013-08-12 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.592 Å) | | 主引用文献 | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
