6P00
 
 | |
6V83
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-1 | | 分子名称: | ACYLATED CEFTAZIDIME, Beta-lactamase | | 著者 | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2019-12-10 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V6G
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | 分子名称: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | 著者 | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2019-12-05 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V6P
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase | | 分子名称: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | 著者 | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2019-12-05 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | 著者 | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | 登録日 | 2020-12-09 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
6V7T
 
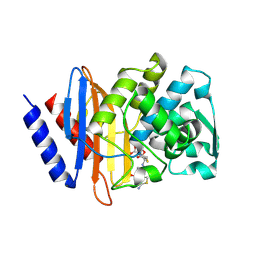 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase in complex with ceftazidime | | 分子名称: | ACYLATED CEFTAZIDIME, Beta-lactamase | | 著者 | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2019-12-09 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
4UUI
 
 | |
8AIS
 
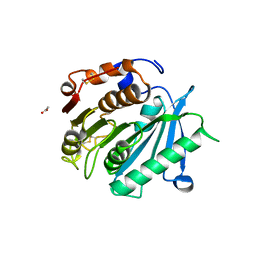 | | Crystal structure of cutinase PsCut from Pseudomonas saudimassiliensis | | 分子名称: | ACETATE ION, Lipase 1 | | 著者 | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | 登録日 | 2022-07-27 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8JIF
 
 | |
7UD4
 
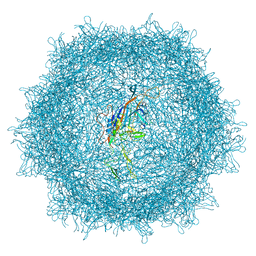 | | Cryo-EM structure of AAV-PHP.eB | | 分子名称: | Capsid protein VP1 | | 著者 | Jang, S, Shen, H.K, Ding, X, Miles, T.F, Gradinaru, V. | | 登録日 | 2022-03-18 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.24 Å) | | 主引用文献 | Structural basis of receptor usage by the engineered capsid AAV-PHP.eB.
Mol Ther Methods Clin Dev, 26, 2022
|
|
6RXQ
 
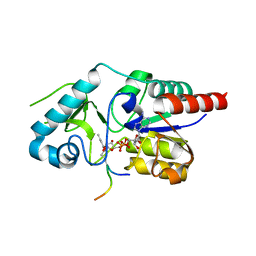 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after soaking | | 分子名称: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4X0K
 
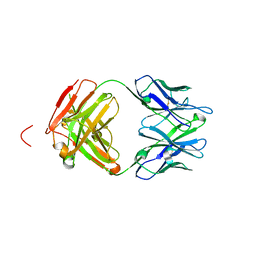 | | Engineered Fab fragment specific for EYMPME (EE) peptide | | 分子名称: | Fab fragment heavy chain, Fab fragment light chain | | 著者 | Johnson, J.L, Lieberman, R.L. | | 登録日 | 2014-11-21 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural and biophysical characterization of an epitope-specific engineered Fab fragment and complexation with membrane proteins: implications for co-crystallization.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6AML
 
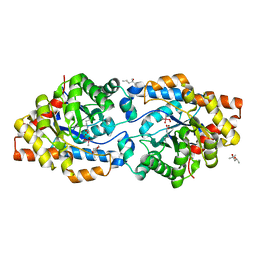 | | Phosphotriesterase variant S8 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | 著者 | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | 登録日 | 2017-08-09 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Phosphotriesterase variant S8
To Be Published
|
|
6RXK
 
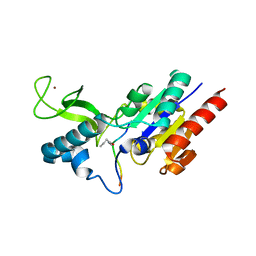 | | Crystal structure of CobB wt in complex with H4K16-Butyryl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXS
 
 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | 分子名称: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXL
 
 | | Crystal structure of CobB wt in complex with H4K16-Crotonyl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXR
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162G) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after co-crystallisation | | 分子名称: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXP
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16-Crotonyl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6B2F
 
 | | Phosphotriesterase variant S5 + TS analogue | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase, ZINC ION, ... | | 著者 | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | 登録日 | 2017-09-20 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.775 Å) | | 主引用文献 | Phosphotriesterase variant S5 + TS analogue
To Be Published
|
|
6RXO
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162A) in complex with H4K16-Buturyl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8P7I
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | (2~{S},6~{R})-2,6-dimethyl-1,3-dioxane-4,4-diol, FORMIC ACID, Parathion hydrolase, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7S
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1,2-ETHANEDIOL, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-31 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7N
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | FORMIC ACID, Parathion hydrolase, ZINC ION | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7V
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1,2-ETHANEDIOL, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | 著者 | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-31 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.737 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7Q
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | 分子名称: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | 著者 | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
