5SVA
 
 | | Mediator-RNA Polymerase II Pre-Initiation Complex | | 分子名称: | 108bp HIS4 Promoter Non-template Strand (-92/+16), 108bp HIS4 Promoter Template Strand (+16/-92), DNA repair helicase RAD25, ... | | 著者 | Robinson, P.J, Bushnell, D.A, Kornberg, R.D. | | 登録日 | 2016-08-05 | | 公開日 | 2016-09-28 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (15.3 Å) | | 主引用文献 | Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex.
Cell, 166, 2016
|
|
5GJR
 
 | | An atomic structure of the human 26S proteasome | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | 登録日 | 2016-07-01 | | 公開日 | 2016-09-07 | | 最終更新日 | 2019-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5J4Z
 
 | | Architecture of tight respirasome | | 分子名称: | COMPLEX I 13KDA/NDUFS6, COMPLEX I 15KDA/NDUFS5, COMPLEX I 18KDA/NDUFS6, ... | | 著者 | Letts, J.A, Fiedorczuk, K, Sazanov, L.A. | | 登録日 | 2016-04-01 | | 公開日 | 2016-09-21 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | The architecture of respiratory supercomplexes.
Nature, 537, 2016
|
|
2JA8
 
 | | CPD lesion containing RNA Polymerase II elongation complex D | | 分子名称: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TP*TTP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*AP*UP)-3', ... | | 著者 | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | 登録日 | 2006-11-23 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
2JA7
 
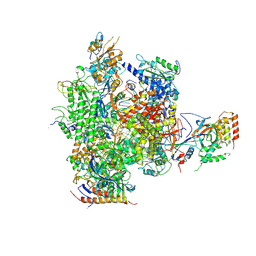 | | CPD lesion containing RNA Polymerase II elongation complex C | | 分子名称: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TTP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | 著者 | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | 登録日 | 2006-11-23 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
2JA6
 
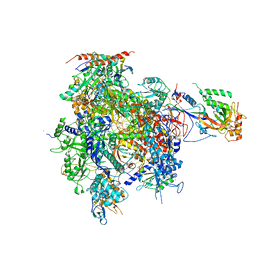 | | CPD lesion containing RNA Polymerase II elongation complex B | | 分子名称: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TTP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | 著者 | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | 登録日 | 2006-11-23 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | CPD damage recognition by transcribing RNA polymerase II.
Science, 315, 2007
|
|
2C2O
 
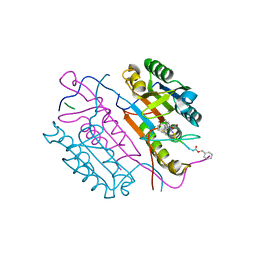 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | 分子名称: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-{4-[BENZYL(METHYL) AMINO]-4-OXOBUTANOYL}-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN-16-OIC ACID, ... | | 著者 | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | 登録日 | 2005-09-29 | | 公開日 | 2006-09-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
8RBX
 
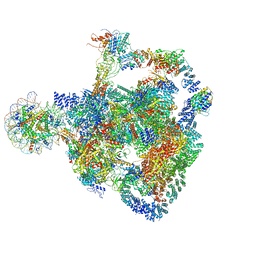 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | 登録日 | 2023-12-05 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
3CWB
 
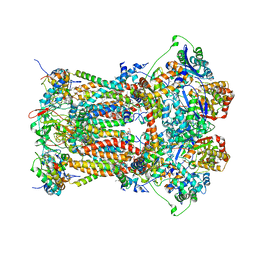 | | Chicken Cytochrome BC1 Complex inhibited by an iodinated analogue of the polyketide Crocacin-D | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, CARDIOLIPIN, ... | | 著者 | Huang, L, Cromartie, T, Viner, R, Crowley, P.J, Berry, E.A. | | 登録日 | 2008-04-21 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D.
Bioorg.Med.Chem., 16, 2008
|
|
6HA8
 
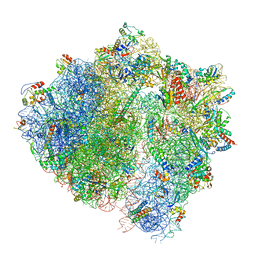 | | Cryo-EM structure of the ABCF protein VmlR bound to the Bacillus subtilis ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | 登録日 | 2018-08-07 | | 公開日 | 2018-08-29 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BZ9
 
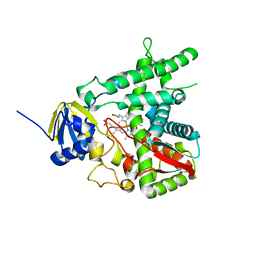 | |
3DGI
 
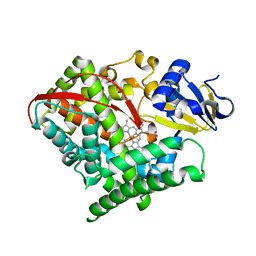 | | Crystal structure of F87A/T268A mutant of CYP BM3 | | 分子名称: | Bifunctional P-450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Le Trong, I, Katayama, J.H, Totah, R.A, Stenkamp, R.E, Fox, E.P. | | 登録日 | 2008-06-13 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Homolytic versus heterolytic dioxygen bond cleavage in cytochrome P450 BM3.
To be Published
|
|
6HA1
 
 | | Cryo-EM structure of a 70S Bacillus subtilis ribosome translating the ErmD leader peptide in complex with telithromycin | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | 登録日 | 2018-08-07 | | 公開日 | 2018-08-29 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ZQG
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-C | | 分子名称: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQD
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
7D63
 
 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-29 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (12.3 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
7D5T
 
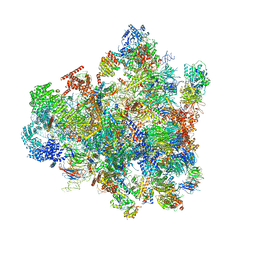 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1)
To Be Published
|
|
6HKO
 
 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | 登録日 | 2018-09-07 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
7AJU
 
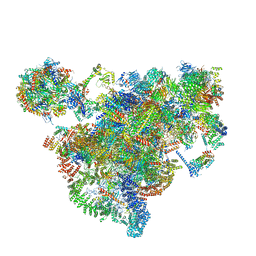 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2020-09-29 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
3EDV
 
 | |
2B63
 
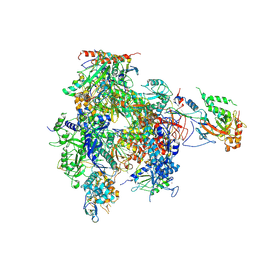 | | Complete RNA Polymerase II-RNA inhibitor complex | | 分子名称: | 31-MER, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | 著者 | Kettenberger, H, Eisenfuehr, A, Brueckner, F, Theis, M, Famulok, M, Cramer, P. | | 登録日 | 2005-09-30 | | 公開日 | 2005-12-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs
Nat.Struct.Mol.Biol., 13, 2006
|
|
2E2J
 
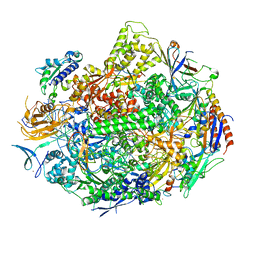 | | RNA polymerase II elongation complex in 5 mM Mg+2 with GMPCPP | | 分子名称: | 27-MER DNA template strand, 5'-D(P*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*A)-3', 5'-R(P*AP*UP*CP*GP*AP*GP*AP*GP*G)-3', ... | | 著者 | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | 登録日 | 2006-11-14 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2E2H
 
 | | RNA polymerase II elongation complex at 5 mM Mg2+ with GTP | | 分子名称: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | 著者 | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | 登録日 | 2006-11-14 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.95 Å) | | 主引用文献 | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2E2I
 
 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dGTP | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', ... | | 著者 | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | 登録日 | 2006-11-14 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2KLE
 
 | |
