3CKI
 
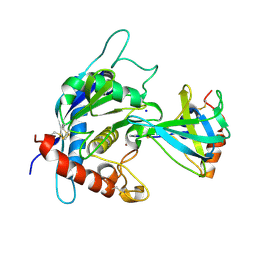 | | Crystal structure of the TACE-N-TIMP-3 complex | | 分子名称: | ADAM 17, Metalloproteinase inhibitor 3, SODIUM ION, ... | | 著者 | Wisniewska, M, Goettig, P, Maskos, K, Belouski, E, Winters, D, Hecht, R, Black, R, Bode, W. | | 登録日 | 2008-03-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex.
J.Mol.Biol., 381, 2008
|
|
3CTL
 
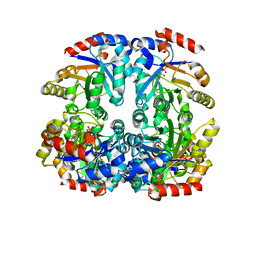 | | Crystal structure of D-Allulose 6-Phosphate 3-Epimerase from Escherichia coli K12 complexed with D-glucitol 6-phosphate and magnesium | | 分子名称: | D-SORBITOL-6-PHOSPHATE, D-allulose-6-phosphate 3-epimerase, MAGNESIUM ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | 登録日 | 2008-04-14 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for substrate specificity in phosphate binding (beta/alpha)8-barrels: D-allulose 6-phosphate 3-epimerase from Escherichia coli K-12.
Biochemistry, 47, 2008
|
|
3CUE
 
 | |
3D21
 
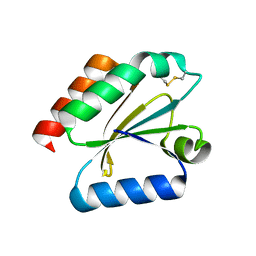 | | Crystal structure of a poplar wild-type thioredoxin h, PtTrxh4 | | 分子名称: | Thioredoxin H-type | | 著者 | Koh, C.S, Didierjean, C, Corbier, C, Rouhier, N, Jacquot, J.P, Gelhaye, E. | | 登録日 | 2008-05-07 | | 公開日 | 2008-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | An Atypical Catalytic Mechanism Involving Three Cysteines of Thioredoxin.
J.Biol.Chem., 283, 2008
|
|
3D5G
 
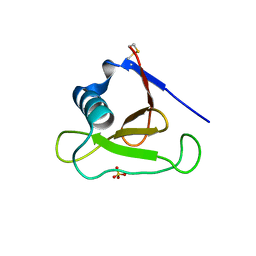 | | Structure of ribonuclease Sa2 complexes with mononucleotides: new aspects of catalytic reaction and substrate recognition | | 分子名称: | Ribonuclease, SULFATE ION | | 著者 | Bauerova-Hlinkova, V, Dvorsky, R, Povazanec, F, Sevcik, J. | | 登録日 | 2008-05-16 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of RNase Sa2 complexes with mononucleotides - new aspects of catalytic reaction and substrate recognition
Febs J., 276, 2009
|
|
3D5S
 
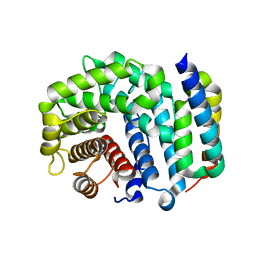 | | Crystal Structure of Efb-C (R131A) / C3d Complex | | 分子名称: | Complement C3, Fibrinogen-binding protein | | 著者 | Geisbrecht, B.V. | | 登録日 | 2008-05-16 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d.
Protein Sci., 17, 2008
|
|
3CTM
 
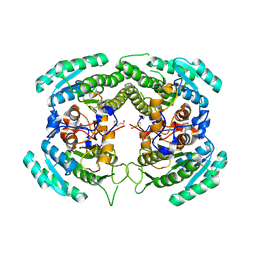 | | Crystal Structure of a Carbonyl Reductase from Candida Parapsilosis with anti-Prelog Stereo-specificity | | 分子名称: | Carbonyl Reductase | | 著者 | Zhang, R, Zhu, G, Li, X, Xu, Y, Zhang, X.C, Rao, Z. | | 登録日 | 2008-04-14 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Protein Sci., 17, 2008
|
|
3DGY
 
 | |
3DH2
 
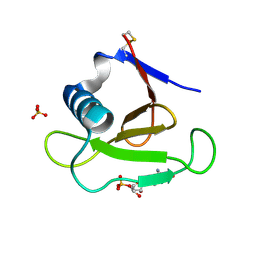 | |
3D92
 
 | | Human carbonic anhydrase II bound with substrate carbon dioxide | | 分子名称: | CARBON DIOXIDE, GLYCEROL, ZINC ION, ... | | 著者 | Domsic, J.F, Avvaru, B.S, McKenna, R. | | 登録日 | 2008-05-26 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Entrapment of carbon dioxide in the active site of carbonic anhydrase II
J.Biol.Chem., 283, 2008
|
|
3DFZ
 
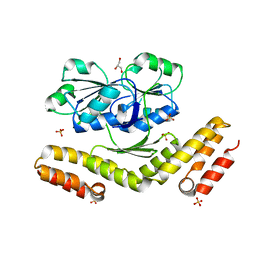 | | SirC, precorrin-2 dehydrogenase | | 分子名称: | GLYCEROL, Precorrin-2 dehydrogenase, SULFATE ION | | 著者 | Schubert, H.L, Hill, C.P, Warren, M.J. | | 登録日 | 2008-06-12 | | 公開日 | 2008-10-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and function of SirC from Bacillus megaterium: a metal-binding precorrin-2 dehydrogenase
Biochem.J., 415, 2008
|
|
3D93
 
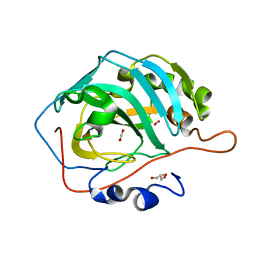 | |
3DA3
 
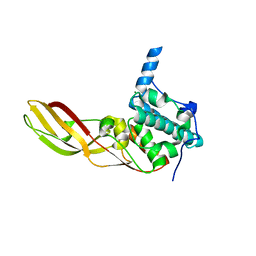 | | Crystal Structure of Colicin M, A Novel Phosphatase Specifically Imported by Escherichia Coli | | 分子名称: | Colicin-M, MAGNESIUM ION | | 著者 | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | 登録日 | 2008-05-28 | | 公開日 | 2008-09-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
6WSK
 
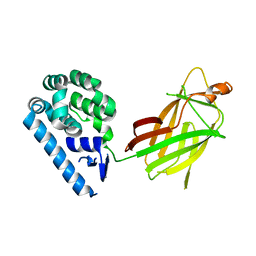 | |
3K9F
 
 | | Detailed structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases | | 分子名称: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA (5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3'), ... | | 著者 | Laponogov, I, Pan, X.-S, Veselkov, D.A, Fisher, L.M, Sanderson, M.R. | | 登録日 | 2009-10-15 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
6L4C
 
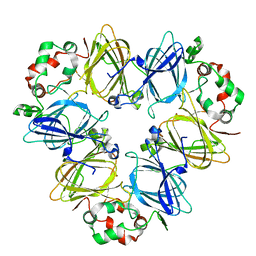 | |
4IML
 
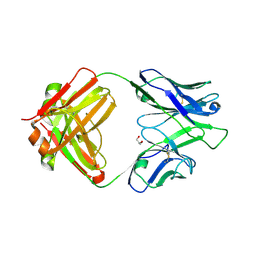 | | CrossFab binding to human Angiopoietin 2 | | 分子名称: | Crossed heavy chain (VH-Ckappa), Crossed light chain (VL-CH1), GLYCEROL | | 著者 | Fenn, S, Schiller, C, Griese, J, Hopfner, K.-P, Kettenberger, H. | | 登録日 | 2013-01-03 | | 公開日 | 2013-04-17 | | 最終更新日 | 2013-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.931 Å) | | 主引用文献 | Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain.
Plos One, 8, 2013
|
|
4ZDM
 
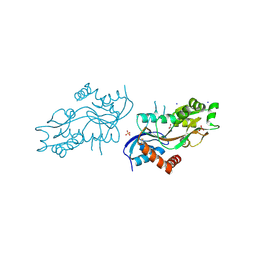 | | Pleurobrachia bachei iGluR3 LBD Glycine Complex | | 分子名称: | GLYCINE, Glutamate receptor kainate-like protein, SODIUM ION, ... | | 著者 | Grey, R.J, Mayer, M.L. | | 登録日 | 2015-04-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6MA6
 
 | |
6MA8
 
 | | Human CYP3A4 bound to PMSF | | 分子名称: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, DIMETHYL SULFOXIDE, ... | | 著者 | Sevrioukova, I.F. | | 登録日 | 2018-08-26 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Interaction of Human Drug-Metabolizing CYP3A4 with Small Inhibitory Molecules.
Biochemistry, 58, 2019
|
|
6MA7
 
 | | Human CYP3A4 bound to an inhibitor fluconazole | | 分子名称: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Cytochrome P450 3A4, ... | | 著者 | Sevrioukova, I.F. | | 登録日 | 2018-08-26 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Interaction of Human Drug-Metabolizing CYP3A4 with Small Inhibitory Molecules.
Biochemistry, 58, 2019
|
|
8Z99
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state +I | | 分子名称: | RNA (49-MER), RNA (54-MER), ZINC ION, ... | | 著者 | Zhang, H, Deng, Z, Li, X. | | 登録日 | 2024-04-22 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8YHE
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state II | | 分子名称: | RNA (29-MER), RNA (46-MER), ZINC ION, ... | | 著者 | Zhang, H, Deng, Z, Li, X. | | 登録日 | 2024-02-28 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8YHD
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state I | | 分子名称: | RNA (35-MER), RNA (53-MER), ZINC ION, ... | | 著者 | Zhang, H, Deng, Z, Li, X. | | 登録日 | 2024-02-28 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4L
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | 分子名称: | RNA (40-MER), RNA (49-MER), ZINC ION, ... | | 著者 | Zhang, H, Deng, Z, Li, X. | | 登録日 | 2024-04-17 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
