5RE9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434836 | | 分子名称: | 2-(4-methylphenoxy)-1-(4-methylpiperazin-4-ium-1-yl)ethanone, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6M5R
 
 | | The coordinates of the apo monomeric terminase complex | | 分子名称: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | 著者 | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | 登録日 | 2020-03-11 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
1TGB
 
 | | CRYSTAL STRUCTURE OF BOVINE TRYPSINOGEN AT 1.8 ANGSTROMS RESOLUTION. II. CRYSTALLOGRAPHIC REFINEMENT, REFINED CRYSTAL STRUCTURE AND COMPARISON WITH BOVINE TRYPSIN | | 分子名称: | CALCIUM ION, TRYPSINOGEN | | 著者 | Bode, W, Fehlhammer, H, Huber, R. | | 登録日 | 1979-03-07 | | 公開日 | 1979-06-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of bovine trypsinogen at 1-8 A resolution. II. Crystallographic refinement, refined crystal structure and comparison with bovine trypsin.
J.Mol.Biol., 111, 1977
|
|
5R8E
 
 | | PanDDA analysis group deposition INTERLEUKIN-1 BETA -- Fragment Z57475877 in complex with INTERLEUKIN-1 BETA | | 分子名称: | Interleukin-1 beta, SULFATE ION, ~{N}-(2-ethyl-1,2,3,4-tetrazol-5-yl)butanamide | | 著者 | De Nicola, G.F, Nichols, C.E. | | 登録日 | 2020-03-03 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
4FSU
 
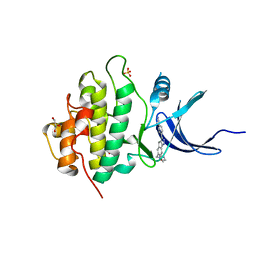 | | Crystal Structure of the CHK1 | | 分子名称: | 4-[6-(1H-imidazol-1-ylmethyl)-7-methoxy-2,4-dihydroindeno[1,2-c]pyrazol-3-yl]benzonitrile, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | 登録日 | 2012-06-27 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the CHK1
To be Published
|
|
7GC3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAG-UCB-cedd89ab-4 (Mpro-x10488) | | 分子名称: | 1-[(2S)-2-(5-cyclopropyl-1,2,4-oxadiazol-3-yl)pyrrolidin-1-yl]-2-(pyridin-3-yl)ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4FTO
 
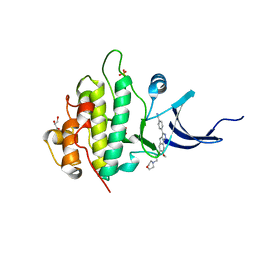 | | Crystal Structure of the CHK1 | | 分子名称: | 4-{7-methoxy-6-[3-(morpholin-4-yl)propoxy]-1,4-dihydroindeno[1,2-c]pyrazol-3-yl}benzonitrile, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | 登録日 | 2012-06-27 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the CHK1
To be Published
|
|
1T7K
 
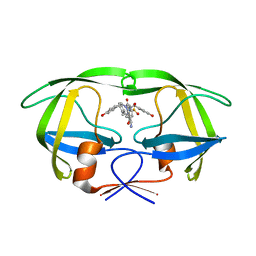 | | Crystal Structure of HIV Protease complexed with Arylsulfonamide azacyclic urea | | 分子名称: | 3-({5-BENZYL-6-HYDROXY-2,4-BIS-(4-HYDROXY-BENZYL)-3-OXO-[1,2,4]-TRIAZEPANE-1-SULFONYL)-BENZONITRILE, Pol polyprotein [Contains: Protease (Retropepsin)] | | 著者 | Huang, P.P, Randolph, J.T, Klein, L.L, Vasavanonda, S, Dekhtyar, T, Stoll, V.S, Kempf, D.J. | | 登録日 | 2004-05-10 | | 公開日 | 2004-10-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synthesis and Antiviral Activity of P1' Arylsulfonamide Azacyclic Urea HIV Protease Inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1OJX
 
 | | Crystal structure of an Archaeal fructose 1,6-bisphosphate aldolase | | 分子名称: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | 著者 | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | 登録日 | 2003-07-16 | | 公開日 | 2003-09-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
4FV9
 
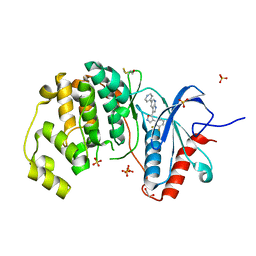 | | Crystal Structure of the ERK2 complexed with E71 | | 分子名称: | 1,2-ETHANEDIOL, 3-[4-(2,3-difluorophenyl)-1,2-oxazol-5-yl]-5-(pyridin-4-yl)-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 1, ... | | 著者 | Kang, Y.N, Stuckey, J.A, Xie, X. | | 登録日 | 2012-06-29 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal Structure of the ERK2 complexed with E71
TO BE PUBLISHED
|
|
1SV3
 
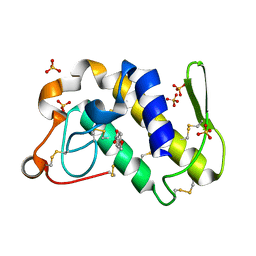 | | Structure of the complex formed between Phospholipase A2 and 4-methoxybenzoic acid at 1.3A resolution. | | 分子名称: | 4-METHOXYBENZOIC ACID, Phospholipase A2, SULFATE ION | | 著者 | Singh, N, Prahathees, E, Jabeen, T, Pal, A, Ethayathulla, A.S, Prem kumar, R, Sharma, S, Singh, T.P. | | 登録日 | 2004-03-27 | | 公開日 | 2004-04-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structures of the complexes of a group IIA phospholipase A2 with two natural anti-inflammatory agents, anisic acid, and atropine reveal a similar mode of binding
Proteins, 64, 2006
|
|
1SVE
 
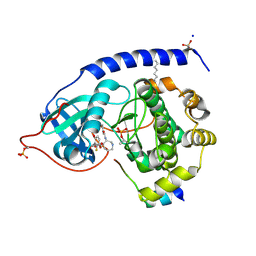 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 1 | | 分子名称: | (4R)-4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZOIC ACID (3R)-3-[(PYRIDINE-4-CARBONYL)AMINO]-AZEPAN-4-YL ESTER, N-OCTANOYL-N-METHYLGLUCAMINE, SODIUM ION, ... | | 著者 | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | 登録日 | 2004-03-29 | | 公開日 | 2005-03-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
4J7E
 
 | | The 1.63A crystal structure of humanized Xenopus MDM2 with a nutlin fragment, RO5524529 | | 分子名称: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-4,5-bis(4-chlorophenyl)-2,4,5-trimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | 著者 | Janson, C, Lukacs, C, Graves, B. | | 登録日 | 2013-02-13 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
2LYI
 
 | |
4FXQ
 
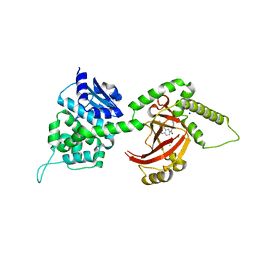 | | Full-length Certhrax toxin from Bacillus cereus in complex with Inhibitor P6 | | 分子名称: | 8-fluoro-2-(3-piperidin-1-ylpropanoyl)-1,3,4,5-tetrahydrobenzo[c][1,6]naphthyridin-6(2H)-one, CHLORIDE ION, Putative ADP-ribosyltransferase Certhrax, ... | | 著者 | Visschedyk, D.D, Dimov, S, Kimber, M.S, Park, H.W, Merrill, A.R. | | 登録日 | 2012-07-03 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9599 Å) | | 主引用文献 | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
3WGU
 
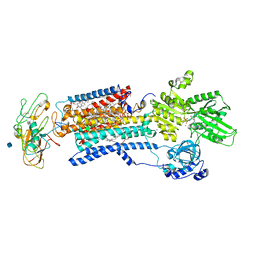 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state without oligomycin | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | 登録日 | 2013-08-09 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
1DKP
 
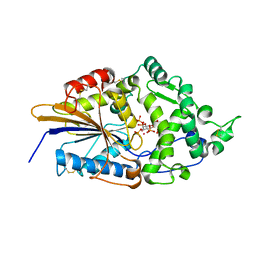 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | 著者 | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | 登録日 | 1999-12-08 | | 公開日 | 2000-08-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
7GB2
 
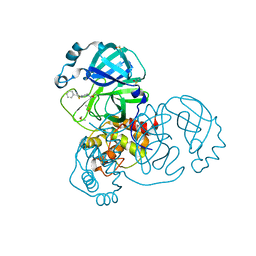 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MED-COV-4280ac29-25 (Mpro-x10155) | | 分子名称: | 1-{4-[(2-benzyl-1,3-thiazol-5-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
1OPJ
 
 | | Structural basis for the auto-inhibition of c-Abl tyrosine kinase | | 分子名称: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, MYRISTIC ACID, ... | | 著者 | Nagar, B, Hantschel, O, Young, M.A, Scheffzek, K, Veach, D, Bornmann, W, Clarkson, B, Superti-Furga, G, Kuriyan, J. | | 登録日 | 2003-03-06 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the autoinhibition of c-Abl tyrosine kinase
Cell(Cambridge,Mass.), 112, 2003
|
|
1SKZ
 
 | | PROTEASE INHIBITOR | | 分子名称: | ANTISTASIN, CHLORIDE ION | | 著者 | Krengel, U, Dijkstra, B.W. | | 登録日 | 1997-04-16 | | 公開日 | 1997-10-22 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-ray structure of antistasin at 1.9 A resolution and its modelled complex with blood coagulation factor Xa.
EMBO J., 16, 1997
|
|
4G1Z
 
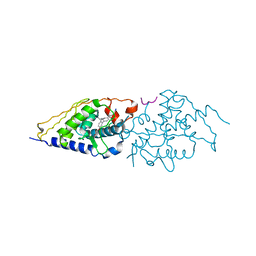 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
3BQ3
 
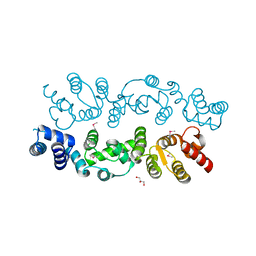 | |
5RYE
 
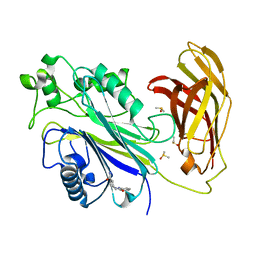 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z33545544 | | 分子名称: | DIMETHYL SULFOXIDE, N~1~-phenylpiperidine-1,4-dicarboxamide, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | 登録日 | 2020-10-30 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RYQ
 
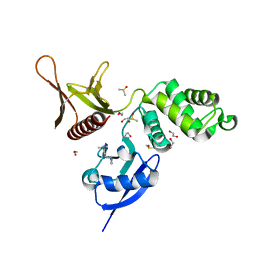 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z57258487 | | 分子名称: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | 著者 | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | 登録日 | 2020-10-30 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
7FTT
 
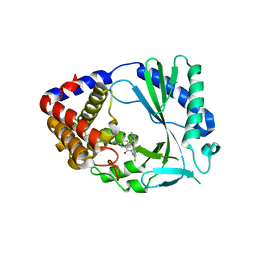 | |
