1WYU
 
 | | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in holo form | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, glycine dehydrogenase (decarboxylating) subunit 1, glycine dehydrogenase subunit 2 (P-protein) | | 著者 | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-02-17 | | 公開日 | 2005-04-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia
Embo J., 24, 2005
|
|
1WUF
 
 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | 分子名称: | MAGNESIUM ION, hypothetical protein lin2664 | | 著者 | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2004-12-07 | | 公開日 | 2004-12-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | 分子名称: | Cold unfolding four helix bundle | | 著者 | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | 登録日 | 2021-11-29 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
5JVG
 
 | | The large ribosomal subunit from Deinococcus radiodurans in complex with avilamycin | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | 著者 | Krupkin, M, Wekselman, I, Matzov, D, Eyal, Z, Diskin Posner, Y, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | 登録日 | 2016-05-11 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.428 Å) | | 主引用文献 | Avilamycin and evernimicin induce structural changes in rProteins uL16 and CTC that enhance the inhibition of A-site tRNA binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1X8H
 
 | |
1PIO
 
 | |
1I9H
 
 | | CORE STREPTAVIDIN-BNA COMPLEX | | 分子名称: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, STREPTAVIDIN | | 著者 | Livnah, O, Huberman, T, Wilchek, M, Bayer, E.A, Eisenberg-Domovich, Y. | | 登録日 | 2001-03-20 | | 公開日 | 2001-09-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Chicken avidin exhibits pseudo-catalytic properties. Biochemical, structural, and electrostatic consequences.
J.Biol.Chem., 276, 2001
|
|
6YM5
 
 | | Crystal structure of BAY-091 with PIP4K2A | | 分子名称: | (2~{R})-2-[[3-cyano-2-[4-(2-fluoranyl-3-methyl-phenyl)phenyl]-1,7-naphthyridin-4-yl]amino]butanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | 著者 | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | 登録日 | 2020-04-07 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM3
 
 | | Crystal structure of Compound 1 with PIP4K2A | | 分子名称: | (2~{R})-2-[[3-cyano-2-[4-(2-ethoxyphenyl)phenyl]-5,8-dihydro-1,7-naphthyridin-4-yl]amino]propanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | 著者 | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | 登録日 | 2020-04-07 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM4
 
 | | Crystal structure of BAY-297 with PIP4K2A | | 分子名称: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | 著者 | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | 登録日 | 2020-04-07 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
1PXE
 
 | |
5N21
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | 分子名称: | 2-[(2~{S})-1-[3-cyano-7-[(2-oxidanylidene-3,4-dihydro-1~{H}-quinolin-6-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl]pyrrolidin-2-yl]ethanoic acid, B-cell lymphoma 6 protein, CHLORIDE ION | | 著者 | Robb, G, Ferguson, A, Hargreaves, D. | | 登録日 | 2017-02-06 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1X
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | 分子名称: | B-cell lymphoma 6 protein, ~{N}-ethyl-5-pyridin-3-yl-pyrazolo[1,5-a]pyrimidin-7-amine | | 著者 | Robb, G, Ferguson, A, Hargreaves, D. | | 登録日 | 2017-02-06 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
2MUB
 
 | |
5N1Z
 
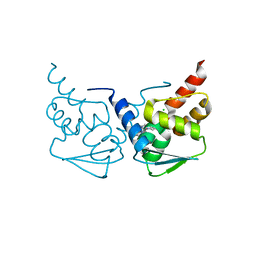 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine macrocyclic ligand | | 分子名称: | B-cell lymphoma 6 protein, CHLORIDE ION, pyrazolo-pyrimidine macrocycle | | 著者 | Robb, G, Ferguson, A, Hargreaves, D. | | 登録日 | 2017-02-06 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
2N81
 
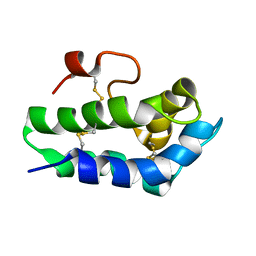 | | Solution Structure of Lipid Transfer Protein From Pea Pisum Sativum | | 分子名称: | Lipid Transfer Protein | | 著者 | Paramonov, A.S, Rumynskiy, E.I, Bogdanov, I.V, Finkina, E.I, Melnikova, D.N, Ovchinnikova, T.V, Shenkarev, Z.O, Arseniev, A.S. | | 登録日 | 2015-09-30 | | 公開日 | 2016-05-11 | | 最終更新日 | 2017-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel lipid transfer protein from the pea Pisum sativum: isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties.
BMC Plant Biol, 16
|
|
5NE0
 
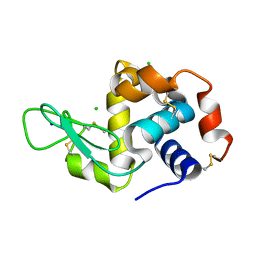 | | Room temperature in-situ structure of hen egg-white lysozyme from crystals enclosed between ultrathin silicon nitride membranes | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Martiel, I, Opara, N, Arnold, S.A, Braun, T, Stahlberg, H, Makita, M, David, C, Padeste, C. | | 登録日 | 2017-03-09 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Direct protein crystallization on ultrathin membranes for diffraction measurements at X-ray free-electron lasers.
J.Appl.Crystallogr., 50, 2017
|
|
2C6W
 
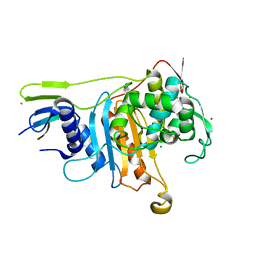 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) FROM STREPTOCOCCUS PNEUMONIAE | | 分子名称: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1A, ZINC ION | | 著者 | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | 登録日 | 2005-11-14 | | 公開日 | 2005-12-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
2MUN
 
 | |
8OLR
 
 | | Structure of yeast 20S proteasome in complex with the natural product beta-lactone inhibitor Cystargolide A | | 分子名称: | CHLORIDE ION, Cystargolide A (bound), MAGNESIUM ION, ... | | 著者 | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | 登録日 | 2023-03-30 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4X6R
 
 | | An Isoform-specific Myristylation Switch Targets RIIb PKA Holoenzymes to Membranes | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Zhang, P, Ye, F, Bastidas, A.C, Kornev, A.P, Ginsberg, M.H, Wu, J, Taylor, S.S. | | 登録日 | 2014-12-09 | | 公開日 | 2015-07-22 | | 最終更新日 | 2020-06-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes.
Structure, 23, 2015
|
|
8OLL
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | 分子名称: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | 著者 | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | 登録日 | 2023-03-30 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4NK9
 
 | | Crystal structure of human fibroblast growth factor receptor 1 kinase domain in complex with pyrazolaminopyrimidine 1 | | 分子名称: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1, N~4~-{5-[2-(3,5-dimethoxyphenyl)ethyl]-1H-pyrazol-3-yl}-N~2~-[(3-methyl-1,2-oxazol-5-yl)methyl]pyrimidine-2,4-diamine, ... | | 著者 | Norman, R.A, Klein, T. | | 登録日 | 2013-11-12 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | FGFR1 Kinase Inhibitors: Close Regioisomers Adopt Divergent Binding Modes and Display Distinct Biophysical Signatures.
ACS Med Chem Lett, 5, 2014
|
|
4NKS
 
 | |
1NOZ
 
 | | T4 DNA POLYMERASE FRAGMENT (RESIDUES 1-388) AT 110K | | 分子名称: | DNA POLYMERASE | | 著者 | Wang, J, Yu, P, Lin, T.C, Konigsberg, W.H, Steitz, T.A. | | 登録日 | 1996-02-16 | | 公開日 | 1996-10-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions.
Biochemistry, 35, 1996
|
|
