3HR3
 
 | |
421D
 
 | |
1QP5
 
 | |
4DOF
 
 | |
4JOM
 
 | | Structure of E. coli Pol III 3mPHP mutant | | 分子名称: | DNA polymerase III subunit alpha, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Barros, T, Guenther, J, Kelch, B, Anaya, J, Prabhakar, A, O'Donnell, M, Kuriyan, J, Lamers, M.H. | | 登録日 | 2013-03-18 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A structural role for the PHP domain in E. coli DNA polymerase III.
Bmc Struct.Biol., 13, 2013
|
|
2K69
 
 | |
2K68
 
 | |
7XFN
 
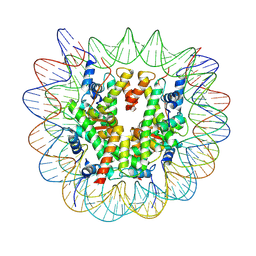 | | Structure of nucleosome-DI complex (-55I, Apo state) | | 分子名称: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Zheng, L, Tsai, B, Gao, N. | | 登録日 | 2022-04-01 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFC
 
 | | Structure of nucleosome-DI complex (-30I, Apo state) | | 分子名称: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Zheng, L, Tsai, B, Gao, N. | | 登録日 | 2022-04-01 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFI
 
 | | Structure of nucleosome-DI complex (-50I, Apo state) | | 分子名称: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Zheng, L, Tsai, B, Gao, N. | | 登録日 | 2022-04-01 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFL
 
 | | Structure of nucleosome-AAG complex (A-53I, free state) | | 分子名称: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Zheng, L, Tsai, B, Gao, N. | | 登録日 | 2022-04-01 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XNP
 
 | | Structure of nucleosome-AAG complex (A-55I, post-catalytic state) | | 分子名称: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Zheng, L, Tsai, B, Gao, N. | | 登録日 | 2022-04-29 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
3FLG
 
 | | The PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 beta | | 分子名称: | DNA (cytosine-5)-methyltransferase 3B | | 著者 | Amaya, M.F, Zeng, H, MacKenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2008-12-18 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 beta
To be Published
|
|
8B0A
 
 | | Cryo-EM structure of ALC1 bound to an asymmetric, site-specifically PARylated nucleosome | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (149-MER) Widom 601 sequence, Histone H2A type 1, ... | | 著者 | Bacic, L, Gaullier, G, Deindl, S. | | 登録日 | 2022-09-07 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Asymmetric nucleosome PARylation at DNA breaks mediates directional nucleosome sliding by ALC1.
Nat Commun, 15, 2024
|
|
2K67
 
 | |
417D
 
 | | A THYMINE-LIKE BASE ANALOGUE FORMS WOBBLE PAIRS WITH ADENINE | | 分子名称: | DNA (5'-D(*CP*AP*CP*GP*(C46)P*G)-3') | | 著者 | Lin, P.K.T, Schuerman, M.H, Moore, G.S, Van Meervelt, L, Loakes, D, Brown, D.M, Moore, M.H. | | 登録日 | 1998-07-15 | | 公開日 | 1998-09-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A thymine-like base analogue forms wobble pairs with adenine in a Z-DNA duplex.
J.Mol.Biol., 282, 1998
|
|
7XVL
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment) | | 分子名称: | DNA (169-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | 著者 | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | 登録日 | 2022-05-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.506 Å) | | 主引用文献 | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment)
To Be Published
|
|
2QSF
 
 | |
4ID3
 
 | |
3QSV
 
 | |
2MWS
 
 | |
2DUN
 
 | |
7PF5
 
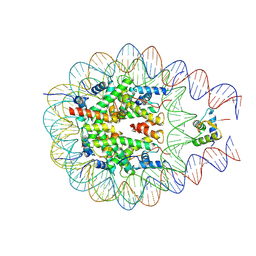 | |
7PF6
 
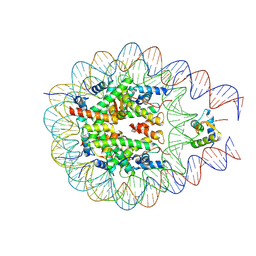 | |
7PF2
 
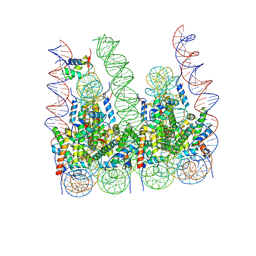 | |
