3VZM
 
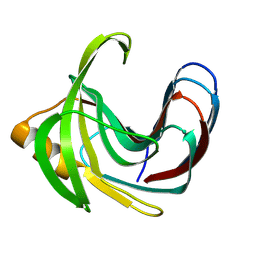 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) E172H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | 分子名称: | Endo-1,4-beta-xylanase, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | 著者 | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | 登録日 | 2012-10-15 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZK
 
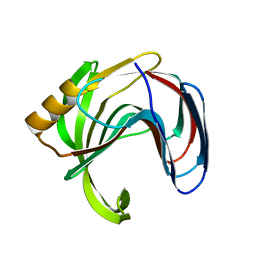 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | 登録日 | 2012-10-14 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZO
 
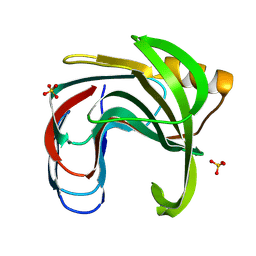 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | 著者 | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | 登録日 | 2012-10-15 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
6UP6
 
 | |
8P2E
 
 | |
2BLM
 
 | |
8PXG
 
 | | Structure of Streptactin, solved at wavelength 2.75 A | | 分子名称: | CHLORIDE ION, GLYCEROL, Streptavidin | | 著者 | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Vecchia, L, Jones, E.Y, Wagner, A. | | 登録日 | 2023-07-23 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
2BOY
 
 | | Crystal structure of 3-ChloroCatechol 1,2-Dioxygenase from Rhodococcus Opacus 1CP | | 分子名称: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 3-CHLOROCATECHOL 1,2-DIOXYGENASE, BENZHYDROXAMIC ACID, ... | | 著者 | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | 登録日 | 2005-04-15 | | 公開日 | 2006-08-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of 3-chlorocatechol 1,2-dioxygenase key enzyme of a new modified ortho-pathway from the Gram-positive Rhodococcus opacus 1CP grown on 2-chlorophenol.
J. Mol. Biol., 360, 2006
|
|
6JMW
 
 | | Structure of the Chromium Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | 分子名称: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, Chromium Protoporphyrin IX, ... | | 著者 | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | 登録日 | 2019-03-13 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JLV
 
 | | Near-Atomic Resolution Structure of the CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | 分子名称: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | 著者 | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | 登録日 | 2019-03-07 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YP7
 
 | | PSII-LHCII C2S2 supercomplex from Pisum sativum grown in high light conditions | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Grinzato, A, Albanese, P, Zanotti, G, Pagliano, C. | | 登録日 | 2020-04-15 | | 公開日 | 2020-11-25 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | High-Light versus Low-Light: Effects on Paired Photosystem II Supercomplex Structural Rearrangement in Pea Plants.
Int J Mol Sci, 21, 2020
|
|
3ZW8
 
 | | Crystal Structure Of Rat Peroxisomal Multifunctional Enzyme Type 1 (rpMFE1) In Apo Form | | 分子名称: | GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, SULFATE ION | | 著者 | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | 登録日 | 2011-07-28 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZWA
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 3S-HYDROXY-HEXANOYL-COA | | 分子名称: | (S)-3-Hydroxyhexanoyl-CoA, GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | 著者 | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | 登録日 | 2011-07-28 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | 登録日 | 2020-03-06 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
3ZWB
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 2TRANS-HEXENOYL-COA | | 分子名称: | (2E)-Hexenoyl-CoA, GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | 著者 | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | 登録日 | 2011-07-28 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZWC
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 3S-HYDROXY-DECANOYL-COA | | 分子名称: | (S)-3-HYDROXYDECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | 登録日 | 2011-07-28 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZW9
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA | | 分子名称: | (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | 著者 | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | 登録日 | 2011-07-28 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
6JMH
 
 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | 分子名称: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | 著者 | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | 登録日 | 2019-03-10 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4AT0
 
 | | The crystal structure of 3-ketosteroid-delta4-(5alpha)-dehydrogenase from Rhodococcus jostii RHA1 | | 分子名称: | 3-KETOSTEROID-DELTA4-5ALPHA-DEHYDROGENASE, ACETATE ION, CHLORIDE ION, ... | | 著者 | van Oosterwijk, N, Knol, J, Dijkhuizen, L, van der Geize, R, Dijkstra, B.W. | | 登録日 | 2012-05-03 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and Catalytic Mechanism of 3-Ketosteroid-{Delta}4-(5Alpha)-Dehydrogenase from Rhodococcus Jostii Rha1 Genome.
J.Biol.Chem., 287, 2012
|
|
1DKM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | 分子名称: | MERCURY (II) ION, PHYTASE | | 著者 | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | 登録日 | 1999-12-08 | | 公開日 | 2000-08-02 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
6JO1
 
 | | Structure of the CYP102A1 Haem Domain with N-(S)-Ibuprofenoyl-L-Phenylalanine | | 分子名称: | (2S)-2-[[(2S)-2-[4-(2-methylpropyl)phenyl]propanoyl]amino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | 著者 | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | 登録日 | 2019-03-19 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6Y3P
 
 | |
6YU9
 
 | |
6YYI
 
 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose | | 分子名称: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | 著者 | Lafite, P, Daniellou, R, Bretagne, D. | | 登録日 | 2020-05-05 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
6JS8
 
 | | Structure of the CYP102A1 Haem Domain with N-Dehydroabietoyl-L-Tryptophan | | 分子名称: | (2S)-2-[[(1R,4aS,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,9,10,10a-hexahydrophenanthren-1-yl]carbonylamino]-3-(1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | 著者 | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | 登録日 | 2019-04-07 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
