2GNX
 
 | | X-ray structure of a hypothetical protein from Mouse Mm.209172 | | 分子名称: | hypothetical protein | | 著者 | Phillips Jr, G.N, McCoy, J.G, Bitto, E, Wesenberg, G.E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-04-11 | | 公開日 | 2006-05-02 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | X-ray structure of a hypothetical protein from Mouse Mm.209172
To be Published
|
|
2H1S
 
 | |
1ZG1
 
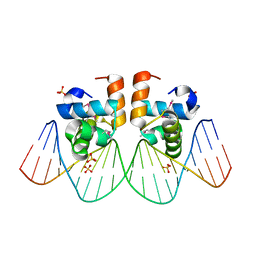 | | NarL complexed to nirB promoter non-palindromic tail-to-tail DNA site | | 分子名称: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*GP*GP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*CP*TP*CP*CP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, ... | | 著者 | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | 登録日 | 2005-04-20 | | 公開日 | 2005-11-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
1G7V
 
 | | CRYSTAL STRUCTURES OF KDO8P SYNTHASE IN ITS BINARY COMPLEXES WITH THE MECHANISM-BASED INHIBITOR | | 分子名称: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, {[(2,2-DIHYDROXY-ETHYL)-(2,3,4,5-TETRAHYDROXY-6-PHOSPHONOOXY-HEXYL)-AMINO]-METHYL}-PHOSPHONIC ACID | | 著者 | Asojo, O.A, Friedman, J.M, Belakhov, V, Shoham, Y, Adir, N, Baasov, T. | | 登録日 | 2000-11-14 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of KDOP synthase in its binary complexes with the substrate phosphoenolpyruvate and with a mechanism-based inhibitor.
Biochemistry, 40, 2001
|
|
1G7U
 
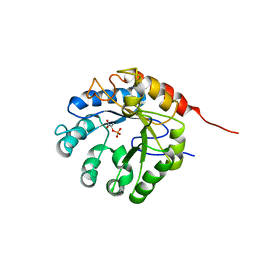 | | CRYSTAL STRUCTURES OF KDO8P SYNTHASE IN ITS BINARY COMPLEX WITH SUBSTRATE PHOSPHOENOL PYRUVATE | | 分子名称: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, PHOSPHOENOLPYRUVATE | | 著者 | Asojo, O.A, Friedman, J.M, Belakhov, V, Shoham, Y, Adir, N, Baasov, T. | | 登録日 | 2000-11-14 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of KDOP synthase in its binary complexes with the substrate phosphoenolpyruvate and with a mechanism-based inhibitor.
Biochemistry, 40, 2001
|
|
2FB4
 
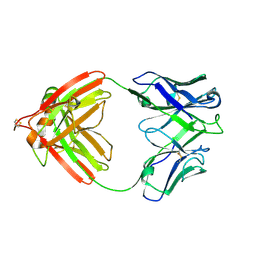 | | DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) | | 分子名称: | IGG1-LAMBDA KOL FAB (HEAVY CHAIN), IGG1-LAMBDA KOL FAB (LIGHT CHAIN) | | 著者 | Marquart, M, Huber, R. | | 登録日 | 1989-04-18 | | 公開日 | 1989-07-12 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I
Biol.Chem.Hoppe-Seyler, 370, 1989
|
|
1BJM
 
 | |
2QXT
 
 | |
4XI8
 
 | |
2QXU
 
 | |
1AI9
 
 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE | | 分子名称: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Whitlow, M, Howard, A.J, Stewart, D. | | 登録日 | 1997-05-01 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
4F0C
 
 | | Crystal structure of the glutathione transferase URE2P5 from Phanerochaete chrysosporium | | 分子名称: | GLYCEROL, Glutathione transferase, OXIDIZED GLUTATHIONE DISULFIDE, ... | | 著者 | Didierjean, C, Favier, F, Roret, T. | | 登録日 | 2012-05-04 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Evolutionary divergence of Ure2pA glutathione transferases in wood degrading fungi.
Fungal Genet Biol, 83, 2015
|
|
1A2D
 
 | | PYRIDOXAMINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | 分子名称: | ADIPOCYTE LIPID BINDING PROTEIN, CHLORIDE ION | | 著者 | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | 登録日 | 1997-12-29 | | 公開日 | 1998-07-01 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
2RFS
 
 | | X-ray structure of SU11274 bound to c-Met | | 分子名称: | Hepatocyte growth factor receptor, N-(3-chlorophenyl)-N-methyl-2-oxo-3-[(3,4,5-trimethyl-1H-pyrrol-2-yl)methyl]-2H-indole-5-sulfonamide | | 著者 | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | 登録日 | 2007-10-01 | | 公開日 | 2007-11-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
1O80
 
 | |
6DPZ
 
 | |
4P5E
 
 | | CRYSTAL STRUCTURE OF HUMAN DNPH1 (RCL) WITH 6-NAPHTHYL-PURINE-RIBOSIDE-MONOPHOSPHATE | | 分子名称: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 6-(naphthalen-2-yl)-9-(5-O-phosphono-beta-D-ribofuranosyl)-9H-purine, CALCIUM ION | | 著者 | Padilla, A, Labesse, G, Kaminski, P.A. | | 登録日 | 2014-03-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: Synthesis, structural studies and cytotoxic activities.
Eur.J.Med.Chem., 85C, 2014
|
|
4AV7
 
 | | Structure determination of the double mutant S233Y F250G from the sec- alkyl sulfatase PisA1 | | 分子名称: | SEC-ALKYLSULFATASE, SULFATE ION, ZINC ION | | 著者 | Knaus, T, Schober, M, Faber, K, Macharaux, P, Wagner, U. | | 登録日 | 2012-05-24 | | 公開日 | 2012-12-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure and Mechanism of an Inverting Alkylsulfatase from Pseudomonas Sp. Dsm6611 Specific for Secondary Alkylsulfates.
FEBS J., 279, 2012
|
|
6DPT
 
 | |
1JFJ
 
 | | NMR SOLUTION STRUCTURE OF AN EF-HAND CALCIUM BINDING PROTEIN FROM ENTAMOEBA HISTOLYTICA | | 分子名称: | CALCIUM-BINDING PROTEIN | | 著者 | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | 登録日 | 2001-06-20 | | 公開日 | 2001-12-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
4Z8B
 
 | | crystal structure of a DGL mutant - H51G H131N | | 分子名称: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, GLYCEROL, ... | | 著者 | Zamora-Caballero, S, Perez, A, Sanz, L, Bravo, J, Calvete, J.J. | | 登録日 | 2015-04-08 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Quaternary structure of Dioclea grandiflora lectin assessed by equilibrium sedimentation and crystallographic analysis of recombinant mutants.
Febs Lett., 589, 2015
|
|
4ZB6
 
 | |
1AZQ
 
 | | HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D BOUND WITH KINKED DNA DUPLEX | | 分子名称: | DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3'), PROTEIN (HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D) | | 著者 | Robinson, H, Gao, Y.-G, Mccrary, B.S, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | 登録日 | 1997-11-20 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Nature, 392, 1998
|
|
3QGH
 
 | |
3QE5
 
 | | Complete structure of Streptococcus mutans Antigen I/II carboxy-terminus | | 分子名称: | CALCIUM ION, MAGNESIUM ION, Major cell-surface adhesin PAc, ... | | 著者 | Larson, M.R, Rajashankar, K.R, Crowley, P.J, Kelly, C, Mitchell, T.J, Brady, L.J, Deivanayagam, C. | | 登録日 | 2011-01-19 | | 公開日 | 2011-04-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the C-terminal Region of Streptococcus mutans Antigen I/II and Characterization of Salivary Agglutinin Adherence Domains.
J.Biol.Chem., 286, 2011
|
|
