2FDC
 
 | |
4R28
 
 | |
346D
 
 | |
2N9H
 
 | | Glucose as a nuclease mimic in DNA | | 分子名称: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | 著者 | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | 登録日 | 2015-11-25 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2H56
 
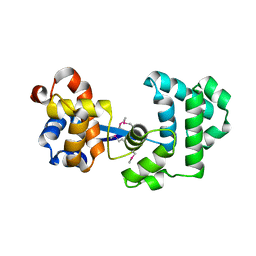 | |
1DA2
 
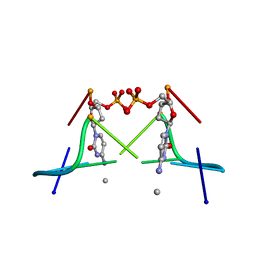 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO4CG): N4-METHOXYCYTOSINE/GUANINE BASE-PAIRS IN Z-DNA | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*(C45)P*G)-3') | | 著者 | Van Meervelt, L, Moore, M.H, Lin, P.K.T, Brown, D.M, Kennard, O. | | 登録日 | 1992-10-17 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular and crystal structure of d(CGCGmo4CG): N4-methoxycytosine.guanine base-pairs in Z-DNA.
J.Mol.Biol., 216, 1990
|
|
7OGG
 
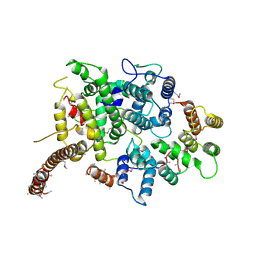 | | Nse5/6 complex | | 分子名称: | DNA repair protein KRE29,DNA repair protein KRE29,DNA repair protein KRE29, Non-structural maintenance of chromosome element 5,Non-structural maintenance of chromosome element 5,Non-structural maintenance of chromosome element 5 | | 著者 | Basquin, J, Taschner, M, Gruber, S. | | 登録日 | 2021-05-06 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-08-11 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Nse5/6 inhibits the Smc5/6 ATPase and modulates DNA substrate binding.
Embo J., 40, 2021
|
|
5ZMD
 
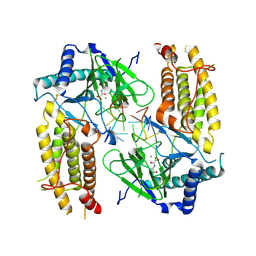 | | Crystal structure of FTO in complex with m6dA modified ssDNA | | 分子名称: | Alpha-ketoglutarate-dependent dioxygenase FTO, DNA (5'-D(P*TP*CP*TP*(6MA)P*TP*AP*TP*CP*G)-3'), MANGANESE (II) ION, ... | | 著者 | Zhang, X, Wei, L.H, Luo, J, Xiao, Y, Liu, J, Zhang, W, Zhang, L, Jia, G.F. | | 登録日 | 2018-04-02 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4IZQ
 
 | |
1D59
 
 | | CRYSTAL STRUCTURE OF 4-STRANDED OXYTRICHA TELOMERIC DNA | | 分子名称: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | 著者 | Kang, C, Zhang, X, Ratliff, R, Moyzis, R, Rich, A. | | 登録日 | 1992-02-25 | | 公開日 | 1993-04-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of four-stranded Oxytricha telomeric DNA.
Nature, 356, 1992
|
|
8KGL
 
 | |
8K0Q
 
 | |
8K0R
 
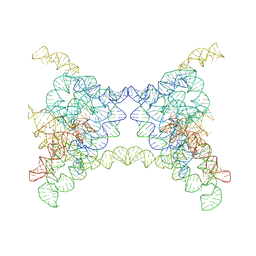 | |
6WQ2
 
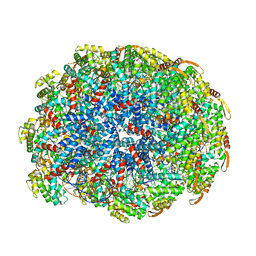 | | Cryo-EM of the S. islandicus filamentous virus, SIFV | | 分子名称: | A-DNA, Structural protein MCP1, Structural protein MCP2 | | 著者 | Wang, F, Baquero, D.P, Su, Z, Zheng, W, Prangishvili, D, Krupovic, M, Egelman, E.H. | | 登録日 | 2020-04-28 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1Q8I
 
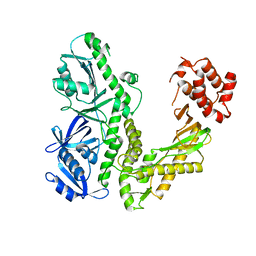 | | Crystal structure of ESCHERICHIA coli DNA Polymerase II | | 分子名称: | DNA polymerase II | | 著者 | Brunzelle, J.S, Muchmore, C.R.A, Mashhoon, N, Blair-Johnson, M, Shuvalova, L, Goodman, M.F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-08-21 | | 公開日 | 2004-01-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Escherichia Coli DNA Polymerase II
To be Published
|
|
2KUZ
 
 | | 2-Aminopurine incorporation perturbs the dynamics and structure of DNA | | 分子名称: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*TP*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*AP*AP*AP*CP*GP*TP*CP*G)-3') | | 著者 | Dallmann, A, Dehmel, L, Peters, T, Muegge, C, Griesinger, C.P, Tuma, J, Ernsting, N.P. | | 登録日 | 2010-03-03 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 2-aminopurine incorporation perturbs the dynamics and structure of DNA.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2KV0
 
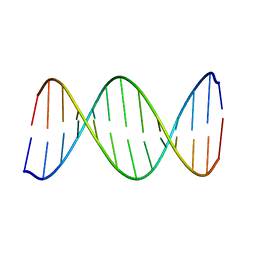 | | 2-Aminopurine incorporation perturbs the dynamics and structure of DNA | | 分子名称: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*TP*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*AP*(2PR)P*AP*CP*GP*TP*CP*G)-3') | | 著者 | Dallmann, A, Dehmel, L, Peters, T, Muegge, C, Griesinger, C.P, Tuma, J, Ernsting, N.P. | | 登録日 | 2010-03-03 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 2-Aminopurine incorporation perturbs the dynamics and structure of DNA.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3P83
 
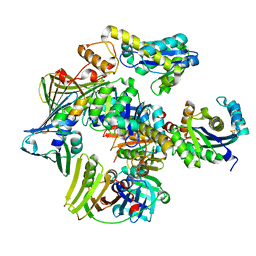 | | Structure of the PCNA:RNase HII complex from Archaeoglobus fulgidus. | | 分子名称: | DNA polymerase sliding clamp, Ribonuclease HII | | 著者 | Bubeck, D, Reijns, M.A, Graham, S.C, Astell, K.R, Jones, E.Y, Jackson, A.P. | | 登録日 | 2010-10-13 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | PCNA directs type 2 RNase H activity on DNA replication and repair substrates.
Nucleic Acids Res., 39, 2011
|
|
2L1P
 
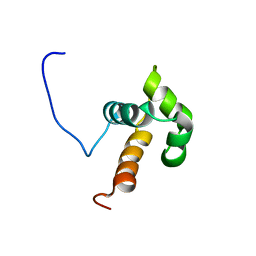 | | NMR solution structure of the N-terminal domain of DNA-binding protein SATB1 from Homo sapiens: Northeast Structural Genomics Target HR4435B(179-250) | | 分子名称: | DNA-binding protein SATB1 | | 著者 | Swapna, G.V.T, Montelione, A.F, Shastry, R, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-08-02 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the N-terminal domain of DNA-binding protein SATB1 from Homo sapiens: Northeast Structural Genomics Target HR4435B(179-250)
To be Published
|
|
4GDF
 
 | |
2MJJ
 
 | |
340D
 
 | |
7KWR
 
 | |
8Q5Q
 
 | |
341D
 
 | |
