9KN7
 
 | | Crystal structure of Horse spleen L-ferritin mutant (Fr-E53F/E56F/E57F/R59L/E60F/E63F) | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | 登録日 | 2024-11-18 | | 公開日 | 2025-03-05 | | 最終更新日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
5X8F
 
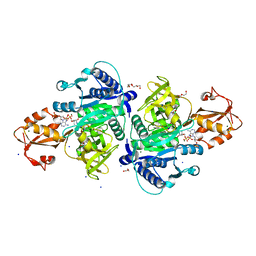 | |
9KRS
 
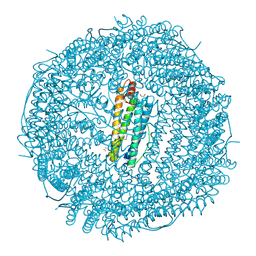 | | Crystal structure of Horse spleen L-ferritin mutant (Fr-E53F/E56F/E57F/R59A/E60F/E63F) | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | 登録日 | 2024-11-28 | | 公開日 | 2025-03-05 | | 最終更新日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KPA
 
 | | Crystal structure of Horse spleen L-ferritin mutant (R59F) | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | 登録日 | 2024-11-22 | | 公開日 | 2025-03-05 | | 最終更新日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
5XFJ
 
 | |
2YIW
 
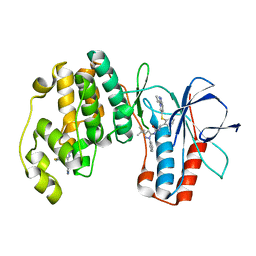 | | triazolopyridine inhibitors of p38 kinase | | 分子名称: | 1-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-3-(2-{[3-(1-methylethyl)[1,2,4]triazolo[4,3-a]pyridin-6-yl]sulfanyl}benzyl)urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | 著者 | Millan, D.S, Anderson, M, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthwaite, R.A, Mahnke, A, Mathais, J.P, Philip, J, Phillips, C, Smith, R.T, Stefamiak, M.H, Yeadon, M. | | 登録日 | 2011-05-17 | | 公開日 | 2011-11-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
5B6S
 
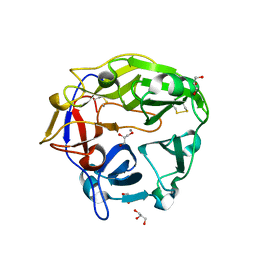 | | Catalytic domain of Coprinopsis cinerea GH62 alpha-L-arabinofuranosidase | | 分子名称: | CALCIUM ION, GLYCEROL, Glycosyl hydrolase family 62 protein | | 著者 | Tonozuka, T. | | 登録日 | 2016-06-01 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the Catalytic Domain of alpha-L-Arabinofuranosidase from Coprinopsis cinerea, CcAbf62A, Provides Insights into Structure-Function Relationships in Glycoside Hydrolase Family 62
Appl. Biochem. Biotechnol., 181, 2017
|
|
5V54
 
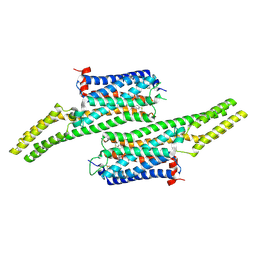 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | 分子名称: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | 著者 | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | 登録日 | 2017-03-13 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
4Q8I
 
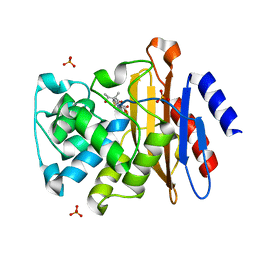 | | Crystal Structure of beta-lactamase from M.tuberculosis covalently complexed with Tebipenem | | 分子名称: | (4R,5S)-3-(1-(4,5-dihydrothiazol-2-yl)azetidin-3-ylthio)-5-((2S,3R)-3-hydroxy-1-oxobutan-2-yl)-4-methyl-4,5- dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Hazra, S, Blanchard, J. | | 登録日 | 2014-04-27 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Tebipenem, a new carbapenem antibiotic, is a slow substrate that inhibits the beta-lactamase from Mycobacterium tuberculosis.
Biochemistry, 53, 2014
|
|
4QFU
 
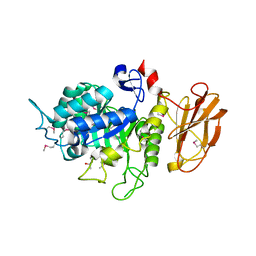 | |
7JGZ
 
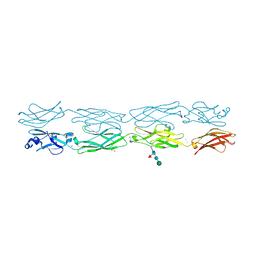 | | Protocadherin gammaC4 EC1-4 crystal structure | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | 登録日 | 2020-07-20 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Elife, 11, 2022
|
|
4RWI
 
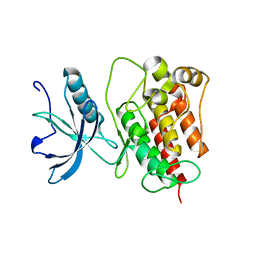 | |
4CYO
 
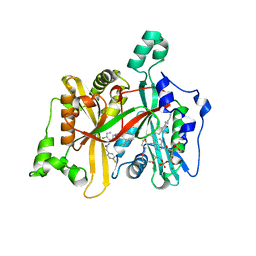 | | Leishmania major N-myristoyltransferase in complex with a hybrid inhibitor (compound 21). | | 分子名称: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, N-{5-[(3S,4R)-1-[(3R)-3-amino-4-(4-chlorophenyl)butanoyl]-4-(hydroxymethyl)pyrrolidin-3-yl]-2-chlorophenyl}-2-(4-fluorophenyl)acetamide, ... | | 著者 | Hutton, J.A, Goncalves, V, Brannigan, J.A, Paape, D, Waugh, T, Roberts, S.M, Bell, A.S, Wilkinson, A.J, Smith, D.F, Leatherbarrow, R.J, Tate, E.W. | | 登録日 | 2014-04-14 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-Based Design of Potent and Selective Leishmania N- Myristoyltransferase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4TMX
 
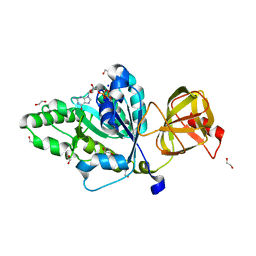 | | Translation initiation factor eIF5B (517-858) mutant D533N from C. thermophilum, bound to GTP and sodium | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Kuhle, B, Ficner, R. | | 登録日 | 2014-06-02 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
5CAP
 
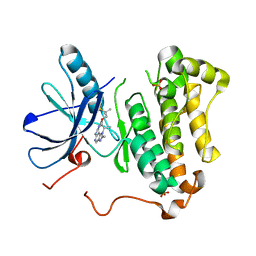 | | EGFR kinase domain mutant "TMLR" with compound 30 | | 分子名称: | 2-methyl-N-[2-(2-methyl-2-methylsulfonyl-propoxy)pyrimidin-4-yl]-1-propan-2-yl-imidazo[4,5-c]pyridin-6-amine, Epidermal growth factor receptor, SULFATE ION | | 著者 | Eigenbrot, C, Yu, C. | | 登録日 | 2015-06-29 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
2Y6U
 
 | |
3KQC
 
 | | Factor xa in complex with the inhibitor 6-(2'- (methylsulfonyl)biphenyl-4-yl)-1-(3-(5-oxo-4,5-dihydro-1h- 1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)-5,6- dihydro-1h-pyrazolo[3,4-c]pyridin-7(4h)-one | | 分子名称: | 6-(2'-(METHYLSULFONYL)BIPHENYL-4-YL)-1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-3-(TRIFLUOROMETHYL)-5,6-DIHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN-7(4H)-ONE, SODIUM ION, factor Xa heavy chain, ... | | 著者 | Sheriff, S. | | 登録日 | 2009-11-17 | | 公開日 | 2010-02-23 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Phenyltriazolinones as potent factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4CGK
 
 | | Crystal structure of the essential protein PcsB from Streptococcus pneumoniae | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Bartual, S.G, Straume, D, Stamsas, G.A, Alfonso, C, Martinez-Ripoll, M, Havarstein, L.S, Hermoso, J.A. | | 登録日 | 2013-11-25 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Basis of Pcsb-Mediated Cell Separation in Streptococcus Pneumoniae.
Nat.Commun., 5, 2014
|
|
6MO2
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | 分子名称: | 1-(4-{5-[(piperidin-4-yl)methoxy]-3-[4-(1H-pyrazol-4-yl)phenyl]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | 著者 | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | 登録日 | 2018-10-03 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
5XM5
 
 | |
7GFJ
 
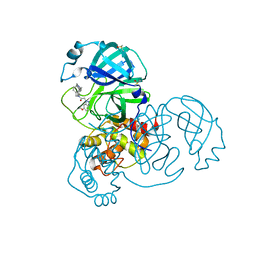 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-c0c213c9-1 (Mpro-x11757) | | 分子名称: | 2-(6-methoxy-1H-benzotriazol-1-yl)-N-[4-(piperidin-4-yl)phenyl]-N-[(pyridin-2-yl)methyl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4RXN
 
 | |
6FPD
 
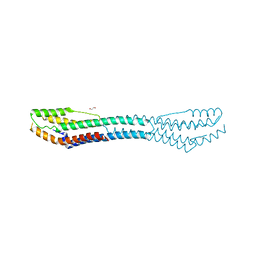 | | AB21 protein from Agaricus bisporus | | 分子名称: | 1,2-ETHANEDIOL, Protein AB21, SODIUM ION | | 著者 | Houser, J, Demo, G, Komarek, J, Wimmerova, M. | | 登録日 | 2018-02-09 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and properties of AB21, a novel Agaricus bisporus protein with structural relation to bacterial pore-forming toxins.
Proteins, 86, 2018
|
|
7GJ2
 
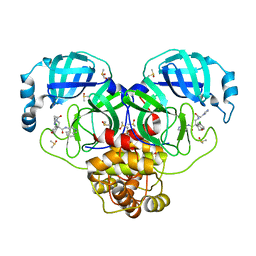 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-70ae9412-1 (Mpro-P0154) | | 分子名称: | (4R)-6-chloro-4-{[2-(1H-imidazol-1-yl)acetamido]methyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGE
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6d04362c-1 (Mpro-x12419) | | 分子名称: | 2-(1H-benzotriazol-1-yl)-N-benzyl-N-[4-(dimethylamino)phenyl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
