5J3F
 
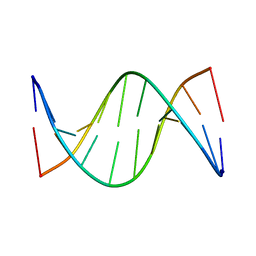 | | NMR solution structure of [Rp, Rp]-PT dsDNA | | 分子名称: | DNA (5'-D(*CP*GP*(RSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(RSG)P*CP*CP*G)-3') | | 著者 | Lan, W, Hu, Z, Cao, C. | | 登録日 | 2016-03-30 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
7K19
 
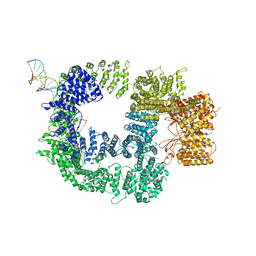 | |
7K1B
 
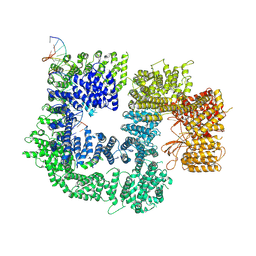 | |
5J3I
 
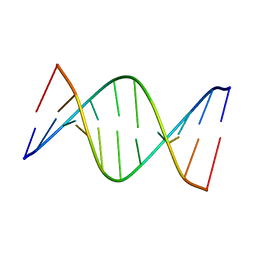 | | NMR solution structure of [Sp, Sp]-PT dsDNA | | 分子名称: | DNA (5'-D(*CP*GP*(SSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(SSG)P*CP*CP*G)-3') | | 著者 | Lan, W, Hu, Z, Cao, C. | | 登録日 | 2016-03-30 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
3MLO
 
 | | DNA binding domain of Early B-cell Factor 1 (Ebf1) bound to DNA (Crystal form I) | | 分子名称: | DNA (5'-D(*CP*TP*TP*TP*AP*TP*TP*CP*CP*CP*AP*TP*GP*GP*GP*AP*AP*TP*AP*AP*AP*G)-3'), Transcription factor COE1, ZINC ION | | 著者 | Treiber, N, Treiber, T, Zocher, G, Grosschedl, R. | | 登録日 | 2010-04-17 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Structure of an Ebf1:DNA complex reveals unusual DNA recognition and structural homology with Rel proteins
Genes Dev., 24, 2010
|
|
3MLN
 
 | | DNA binding domain of Early B-cell Factor 1 (Ebf1) bound to DNA (crystal form II) | | 分子名称: | DNA (5'-D(*CP*TP*TP*TP*AP*TP*TP*CP*CP*CP*AP*TP*GP*GP*GP*AP*AP*TP*AP*AP*AP*G)-3'), Transcription factor COE1, ZINC ION | | 著者 | Treiber, N, Treiber, T, Zocher, G, Grosschedl, R. | | 登録日 | 2010-04-17 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of an Ebf1:DNA complex reveals unusual DNA recognition and structural homology with Rel proteins
Genes Dev., 24, 2010
|
|
5J3G
 
 | |
7K10
 
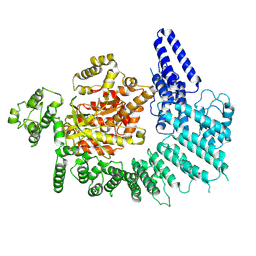 | |
2UUG
 
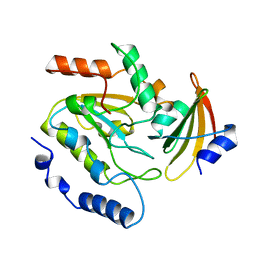 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH H187D MUTANT UDG AND WILD-TYPE UGI | | 分子名称: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | 登録日 | 1998-10-31 | | 公開日 | 1999-03-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
4EQ5
 
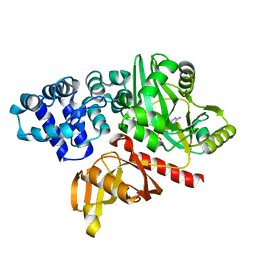 | | DNA ligase from the archaeon Thermococcus sibiricus | | 分子名称: | ADENOSINE MONOPHOSPHATE, DNA ligase | | 著者 | Petrova, T, Bezsudnova, E.Y, Dorokhov, B.D, Slutskaya, E.S, Polyakov, K.M, Dorovatovskiy, P.V, Ravin, N.V, Skryabin, K.G, Kovalchuk, M.V, Popov, V.O. | | 登録日 | 2012-04-18 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Expression, purification, crystallization and preliminary crystallographic analysis of a thermostable DNA ligase from the archaeon Thermococcus sibiricus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1UUG
 
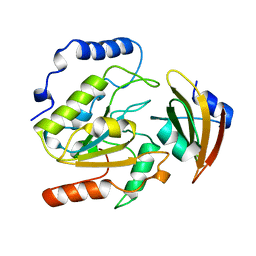 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | 分子名称: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | 登録日 | 1998-10-31 | | 公開日 | 1999-03-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
8VDT
 
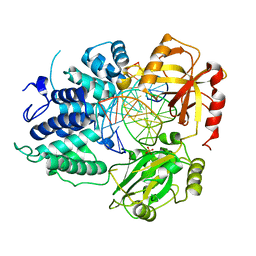 | | DNA Ligase 1 with nick DNA 3'rA:T | | 分子名称: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*TP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*A)-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | 著者 | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | 登録日 | 2023-12-17 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VDS
 
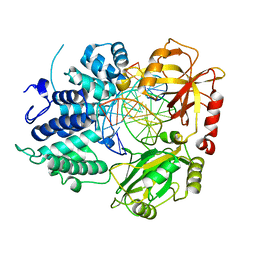 | | DNA Ligase 1 with nick DNA 3'rG:C | | 分子名称: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*G)-D(P*GP*TP*CP*GP*GP*AP*C)-3') | | 著者 | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | 登録日 | 2023-12-17 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
7K17
 
 | |
6DBL
 
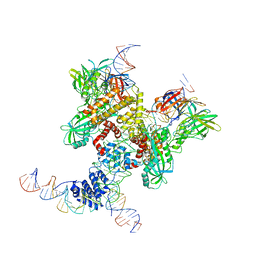 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (5.001 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3W1O
 
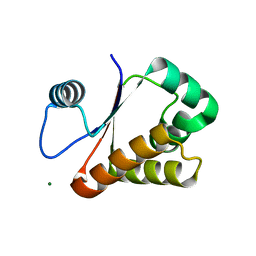 | | Neisseria DNA mimic protein DMP12 | | 分子名称: | DNA mimic protein DMP12, MAGNESIUM ION | | 著者 | Wang, H.C, Ko, T.P, Wu, M.L, Wang, A.H.J. | | 登録日 | 2012-11-19 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein
Nucleic Acids Res., 41, 2013
|
|
6DBQ
 
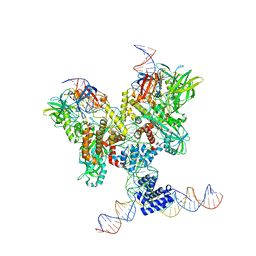 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.22 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBT
 
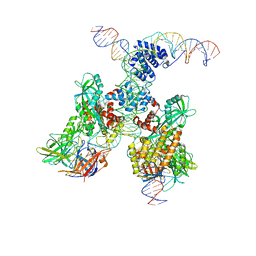 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7CC9
 
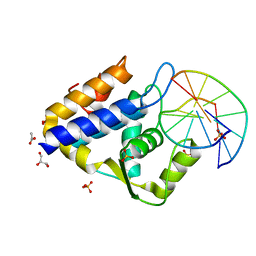 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | 分子名称: | ACETATE ION, DNA (5'-D(*GP*GP*CP*GP*GS*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*CP*GP*CP*C)-3'), ... | | 著者 | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | 登録日 | 2020-06-16 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.063 Å) | | 主引用文献 | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
5WFY
 
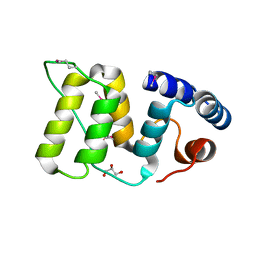 | |
6DBU
 
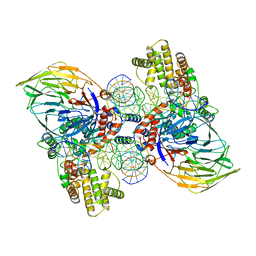 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBV
 
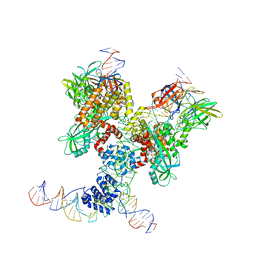 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.291 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBO
 
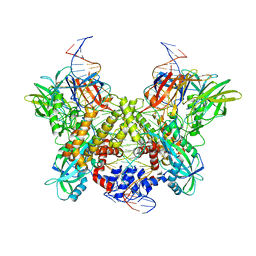 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6W0Q
 
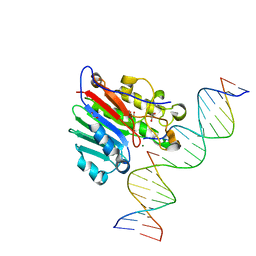 | | APE1 endonuclease product complex D148E | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | 著者 | Freudenthal, B.D, Whitaker, A.M. | | 登録日 | 2020-03-02 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W43
 
 | |
