9FA3
 
 | | Gcase in complex with small molecule inhibitor 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | 著者 | Tisi, D, Cleasby, A. | | 登録日 | 2024-05-10 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
9FA6
 
 | | Gcase in complex with small molecule inhibitor 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | 著者 | Tisi, D, Cleasby, A. | | 登録日 | 2024-05-10 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
2RF0
 
 | | Crystal structure of human mixed lineage kinase MAP3K10 SH3 domain | | 分子名称: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase 10 | | 著者 | Ugochukwu, E, Eswaran, J, Elkins, J, Keates, T, Pike, A.C.W, Berridge, G, Savitsky, P, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2007-09-27 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of human Mixed lineage kinase MAP3K10 SH3 domain.
To be Published
|
|
1BDG
 
 | |
5XLY
 
 | | Crystal structure of CheR1 in complex with c-di-GMP-bound MapZ | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608 | | 著者 | Yuan, Z, Zhu, Y, Gu, L. | | 登録日 | 2017-05-12 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.763 Å) | | 主引用文献 | Structural basis for the regulation of chemotaxis by MapZ in the presence of c-di-GMP
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6T6G
 
 | | Bacteroides salyersiae GH164 beta-mannosidase in complex with noeuromycin | | 分子名称: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Armstrong, Z, Davies, G. | | 登録日 | 2019-10-18 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structure and function ofBs164 beta-mannosidase fromBacteroides salyersiaethe founding member of glycoside hydrolase family GH164.
J.Biol.Chem., 295, 2020
|
|
4LE7
 
 | | The Crystal Structure of Pyocin L1 at 2.09 Angstroms | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyocin L1 | | 著者 | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, R.J, Walker, D. | | 登録日 | 2013-06-25 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
2XVN
 
 | | A. fumigatus chitinase A1 phenyl-methylguanylurea complex | | 分子名称: | 1-METHYL-3-(N-PHENYLCARBAMIMIDOYL)UREA, ASPERGILLUS FUMIGATUS CHITINASE A1, CHLORIDE ION | | 著者 | Rush, C.L, Schuttelkopf, A.W, Hurtado-Guerrero, R, Blair, D.E, Ibrahim, A.F.M, Desvergnes, S, Eggleston, I.M, van Aalten, D.M.F. | | 登録日 | 2010-10-26 | | 公開日 | 2010-11-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Natural Product-Guided Discovery of a Fungal Chitinase Inhibitor.
Chem.Biol., 17, 2010
|
|
6UUO
 
 | | Crystal structure of BRAF kinase domain bound to the PROTAC P4B | | 分子名称: | N-(3-{5-[(1-acetylpiperidin-4-yl)(methyl)amino]-3-(pyrimidin-5-yl)-1H-pyrrolo[3,2-b]pyridin-1-yl}-2,4-difluorophenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | 著者 | Maisonneuve, P, Posternak, G, Kurinov, I, Sicheri, F. | | 登録日 | 2019-10-30 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.288 Å) | | 主引用文献 | Functional characterization of a PROTAC directed against BRAF mutant V600E.
Nat.Chem.Biol., 16, 2020
|
|
5X5G
 
 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with OP0595 | | 分子名称: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, SODIUM ION, ... | | 著者 | Wachino, J, Jin, W, Arakawa, Y. | | 登録日 | 2017-02-15 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
7GI0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-92e193ae-1 (Mpro-P0034) | | 分子名称: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5EIK
 
 | | Structure of a Trimeric Intracellular Cation channel from C. elegans in the absence of Ca2+ | | 分子名称: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | 著者 | Hu, M.H, Yang, H.T, Liu, Z.F. | | 登録日 | 2015-10-30 | | 公開日 | 2016-10-05 | | 最終更新日 | 2025-09-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Pore architecture of TRIC channels and insights into their gating mechanism.
Nature, 538, 2016
|
|
6LY6
 
 | | PylRS C-terminus domain mutant bound with 3-(1-Naphthyl)-L-alanine and AMPNP | | 分子名称: | MAGNESIUM ION, NAPHTHALEN-2-YL-3-ALANINE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Weng, J.H, Tsai, M.D, Wang, Y.S. | | 登録日 | 2020-02-13 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5001328 Å) | | 主引用文献 | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6DIU
 
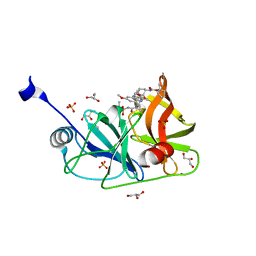 | | Crystal structure of HCV NS3/4A protease in complex with P4-3(AJ-74) | | 分子名称: | 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, GLYCEROL, NS3 protease, ... | | 著者 | Matthew, A.N, Schiffer, C.A. | | 登録日 | 2018-05-23 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.868 Å) | | 主引用文献 | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
7GIY
 
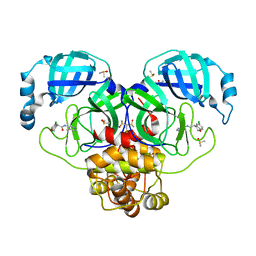 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BRU-CON-c4e3408a-1 (Mpro-P0145) | | 分子名称: | (4R)-6-chloro-N-(4-methylpyridin-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6C57
 
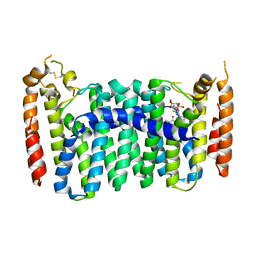 | | Crystal structure of mutant human geranylgeranyl pyrophosphate synthase (Y246D) in complex with bisphosphonate inhibitor FV0109 | | 分子名称: | Geranylgeranyl pyrophosphate synthase, {[(2-{3-[(4-fluorobenzene-1-carbonyl)amino]phenyl}thieno[2,3-d]pyrimidin-4-yl)amino]methylene}bis(phosphonic acid) | | 著者 | Park, J, Bin, X, Vincent, F, Tsantrizos, Y.S, Berghuis, A.M. | | 登録日 | 2018-01-15 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Unraveling the Prenylation-Cancer Paradox in Multiple Myeloma with Novel Geranylgeranyl Pyrophosphate Synthase (GGPPS) Inhibitors.
J. Med. Chem., 61, 2018
|
|
3EKU
 
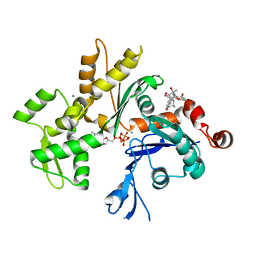 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | 分子名称: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | 著者 | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | 登録日 | 2008-09-19 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
7GL7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-6bf93aa8-1 (Mpro-P1783) | | 分子名称: | (4S)-6-chloro-4-methoxy-N-[7-(methylsulfamoyl)isoquinolin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2Q3G
 
 | | Structure of the PDZ domain of human PDLIM7 bound to a C-terminal extension from human beta-tropomyosin | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ and LIM domain protein 7 | | 著者 | Gileadi, C, Papagrigoriou, E, Elkins, J, Burgess-Brown, N, Salah, E, Gileadi, O, Umeano, C, Bunkoczi, G, von Delft, F, Uppenberg, J, Pike, A.C.W, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | 登録日 | 2007-05-30 | | 公開日 | 2007-06-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
9NBS
 
 | | Structure of HPK1 with compound R2 | | 分子名称: | (1R,3S,5'P)-4'-chloro-3-methyl-5'-[(3M)-3-(1-methyl-2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1',2'-dihydrospiro[cyclopentane-1,3'-pyrrolo[2,3-b]pyridine]-3-carboxamide, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Kiefer, J.R, Walters, B.T, Wang, W, Wu, P, Duo, Y. | | 登録日 | 2025-02-14 | | 公開日 | 2025-06-25 | | 最終更新日 | 2025-07-02 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Integrating Hydrogen Exchange with Molecular Dynamics for Improved Ligand Binding Predictions.
J.Chem.Inf.Model., 65, 2025
|
|
5AND
 
 | | Crystal structure of CDK2 in complex with 2-imidazol-1-yl-1H- benzimidazole processed with the CrystalDirect automated mounting and cryo-cooling technology | | 分子名称: | 2-IMIDAZOL-1-YL-1H-BENZIMIDAZOLE, CYCLIN-DEPENDENT KINASE 2 | | 著者 | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | 登録日 | 2015-09-07 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7GF2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MIC-UNK-08cd9c58-1 (Mpro-x11548) | | 分子名称: | 2-(3-chlorophenyl)-N-(1,7-naphthyridin-5-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7KL0
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) | | 分子名称: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | 著者 | Ozden, C, Stratton, M.M, Garman, S.C. | | 登録日 | 2020-10-28 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
5XKR
 
 | | Crystal structure of Msmeg3575 in complex with benzoguanamine | | 分子名称: | 6-phenyl-1,3,5-triazine-2,4-diamine, ACETATE ION, CMP/dCMP deaminase, ... | | 著者 | Gaded, V.M, Anand, R. | | 登録日 | 2017-05-09 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Selective Deamination of Mutagens by a Mycobacterial Enzyme
J. Am. Chem. Soc., 139, 2017
|
|
7GGN
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ERI-UCB-d6de1f3c-1 (Mpro-x12679) | | 分子名称: | 3C-like proteinase, 4-[4-(3-chlorophenyl)-3-oxopiperazine-1-carbonyl]quinolin-2(1H)-one, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.928 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
