2KCF
 
 | |
5NTN
 
 | | Structural states of RORgt: X-ray elucidation of molecular mechanisms and binding interactions for natural and synthetic compounds | | 分子名称: | (5R,10S,13R,14R,17R)-17-((R,E)-7-hydroxy-6-methylhept-5-en-2-yl)-4,4,10,13,14-pentamethyl-1,2,5,6,10,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthrene-3,7(4H)-dione, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | 著者 | Kallen, J. | | 登録日 | 2017-04-28 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural States of ROR gamma t: X-ray Elucidation of Molecular Mechanisms and Binding Interactions for Natural and Synthetic Compounds.
ChemMedChem, 12, 2017
|
|
2KBU
 
 | |
7LVS
 
 | | The CBP TAZ1 Domain in Complex with a CITED2-HIF-1-Alpha Fusion Peptide | | 分子名称: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone lysine acetyltransferase CREBBP, ZINC ION | | 著者 | Appling, F.D, Berlow, R.B, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | 登録日 | 2021-02-26 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | The molecular basis of allostery in a facilitated dissociation process.
Structure, 29, 2021
|
|
3P8O
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with des-bromine analogue of BI 201335 | | 分子名称: | HCV non-structural protein 4A, HCV serine protease NS3, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-4-[(7-methoxy-2-{2-[(2-methylpropanoyl)amino]-1,3-thiazol-4-yl}quinolin-4-yl)oxy]-L-prolinamide, ... | | 著者 | Lemke, C.T. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-26 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335.
J.Biol.Chem., 286, 2011
|
|
3UA1
 
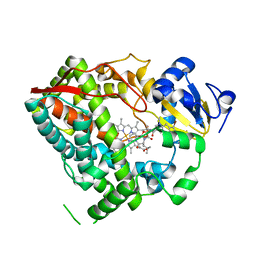 | |
3AW7
 
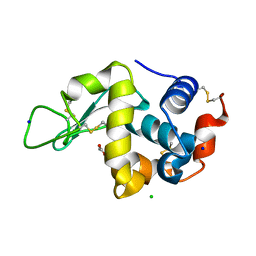 | |
3P8N
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with BI 201335 | | 分子名称: | HCV non-structural protein 4A, HCV serine protease NS3, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-4-[(8-bromo-7-methoxy-2-{2-[(2-methylpropanoyl)amino]-1,3-thiazol-4-yl}quinolin-4-yl)oxy]-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-L-prolinamide, ... | | 著者 | Lemke, C.T. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-26 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335.
J.Biol.Chem., 286, 2011
|
|
3OJ8
 
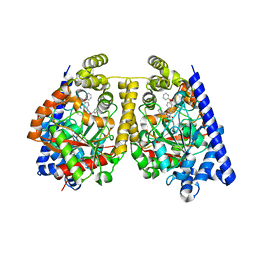 | | Alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints in the Acyl Side Chain | | 分子名称: | (S)-[(2S)-6-phenoxy-1,2,3,4-tetrahydronaphthalen-2-yl](5-pyridin-2-yl-1,3-oxazol-2-yl)methanol, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | 著者 | Mileni, M, Stevens, R.C, Boger, D.L. | | 登録日 | 2010-08-20 | | 公開日 | 2011-07-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints in the Acyl Side Chain
J.Med.Chem., 54, 2011
|
|
2N9E
 
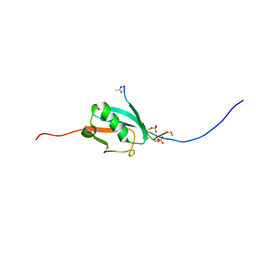 | |
2JX8
 
 | | Solution structure of hPCIF1 WW domain | | 分子名称: | Phosphorylated CTD-interacting factor 1 | | 著者 | Kouno, T, Iwamoto, Y, Hirose, Y, Aizawa, T, Demura, M, Kawano, K, Ohkuma, Y, Mizuguchi, M. | | 登録日 | 2007-11-09 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1H, 13C, and 15N resonance assignments of hPCIF1 WW domain
To be Published
|
|
2LH0
 
 | |
4XKH
 
 | | CRYSTAL STRUCTURE OF THE AIRAPL TANDEM UIMS IN COMPLEX WITH A LYS48-LINKED TRI-UBIQUITIN | | 分子名称: | AN1-type zinc finger protein 2B, Polyubiquitin-C | | 著者 | Rahighi, S, Kawasaki, M, Stanhill, A, Wakatsuki, S. | | 登録日 | 2015-01-11 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains.
Structure, 24, 2016
|
|
4YCU
 
 | | Crystal structure of cladosporin in complex with human lysyl-tRNA synthetase | | 分子名称: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, GLYCEROL, LYSINE, ... | | 著者 | Fang, P, Wang, J, Guo, M. | | 登録日 | 2015-02-20 | | 公開日 | 2015-06-10 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Specific Inhibition of tRNA Synthetase by an ATP Competitive Inhibitor.
Chem. Biol., 22, 2015
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | 分子名称: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | 登録日 | 2020-05-16 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
4YCW
 
 | | Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant | | 分子名称: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, LYSINE, Lysine--tRNA ligase, ... | | 著者 | Fang, P, Wang, J, Guo, M. | | 登録日 | 2015-02-20 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis for Specific Inhibition of tRNA Synthetase by ATP Competitive Inhibitor
Chem.Biol., 22, 2015
|
|
3C7O
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with cellotetraose. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-08 | | 公開日 | 2008-11-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7F
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-07 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3DFX
 
 | | Opposite GATA DNA binding | | 分子名称: | DNA (5'-D(*DAP*DAP*DGP*DGP*DTP*DTP*DAP*DTP*DCP*DTP*DCP*DTP*DGP*DAP*DTP*DTP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DTP*DGP*DAP*DTP*DAP*DAP*DAP*DTP*DCP*DAP*DGP*DAP*DGP*DAP*DTP*DAP*DAP*DCP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | 著者 | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | 登録日 | 2008-06-12 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
3C7H
 
 | | Crystal structure of glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with AXOS-4-0.5. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-07 | | 公開日 | 2008-11-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
1FBI
 
 | |
3C7G
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with xylotetraose. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-07 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7E
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-07 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
1H6V
 
 | | Mammalian thioredoxin reductase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOREDOXIN REDUCTASE | | 著者 | Sandalova, T, Zhong, L, Lindqvist, Y, Holmgren, A, Schneider, G. | | 登録日 | 2001-06-27 | | 公開日 | 2001-08-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Three-Dimensional Structure of a Mammalian Thioredoxin Reductase: Implication for Mechanism and Evolution of a Selenocysteine Dependent Enzyme
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6OJW
 
 | | Crystal structure of Sphingomonas paucimobilis TMY1009 holo-LsdA | | 分子名称: | FE (III) ION, GLYCEROL, Lignostilbene-alpha,beta-dioxygenase isozyme I, ... | | 著者 | Kuatsjah, E, Verstraete, M.M, Kobylarz, M.J, Liu, A.K.N, Murphy, M.E.P, Eltis, L.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase.
J.Biol.Chem., 294, 2019
|
|
